Summary
Highly enriched germinal center (GC) B cell populations are essential for studying humoral immunity. Current MACS protocols that isolate untouched GC B cells require GC induction and typically require further FACS purification with direct antibody labeling to achieve sufficiently high purities. We present a MACS protocol with progressive and repeated negative selections that yields highly purified untouched GC B cells from both unimmunized and GC-induced mice and allows further FACS isolation of unlabeled GC B cells from remaining debris by scatter.
Subject areas: Cell Biology, Cell isolation, Immunology, Model Organisms
Graphical abstract
Highlights
-
•
Isolates highly enriched untouched GC B cells from both naive and immunized mice
-
•
Increases GC B cell purities by progressive and repeated negative selection steps
-
•
Minimizes cell loss by diminishing DNA-mediated nonspecific adherence to beads
-
•
Enables FACS isolation of unlabeled GC B cells by scatter for removing cell debris
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Highly enriched germinal center (GC) B cell populations are essential for studying humoral immunity. Current MACS protocols that isolate untouched GC B cells require GC induction and typically require further FACS purification with direct antibody labeling to achieve sufficiently high purities. We present a MACS protocol with progressive and repeated negative selections that yields highly purified untouched GC B cells from both unimmunized and GC-induced mice and allows further FACS isolation of unlabeled GC B cells from remaining debris by scatter.
Before you begin
Fluorescence-activated cell sorting (FACS) remains a standard method for isolating highly enriched mouse GC B cells. However, using FACS to isolate large numbers of GC B cells is time-consuming and causes cell stress. Furthermore, while forward and side scatter can identify cells by cell size and granularity respectively, scatter cannot be used to discriminate GC B cells from other cell types. For FACS, GC B cells need to be labeled by fluorescent conjugated antibodies, such as anti-Fas antibody, which may impair cell activity and viability (Goillot et al., 1997; Wise et al., 2013).
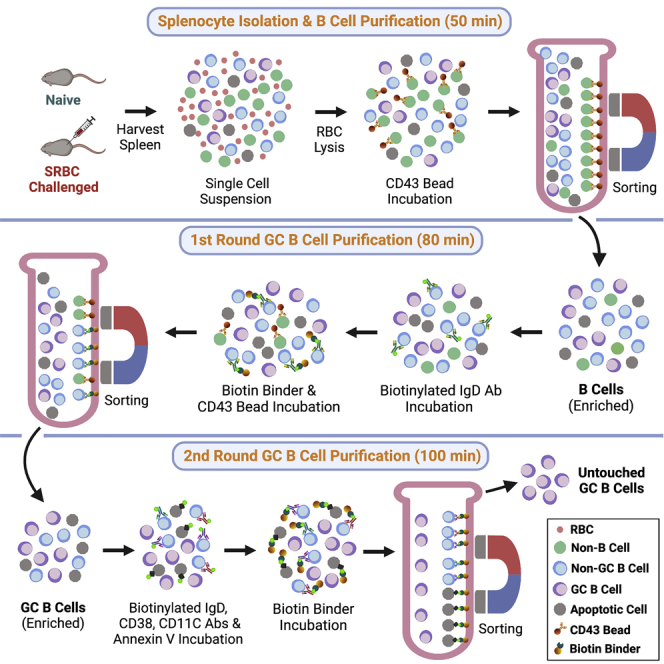
Unlike FACS, magnetic-activated cell sorting (MACS) provides a means for fast bulk isolation of unlabeled GC B cells from mouse spleens by negative selection of antibody-labeled non-GC splenocytes. However, current MACS protocols cannot be used for mice that have limited GC formation because they require sheep red blood cell (SRBC) challenge to induce robust GC responses. Therefore, the use of current MACS protocols is limited if GC formation is irresponsive to SRBC challenge due to gene mutations, or SRBC challenge affects the intended downstream studies. In addition, compared to FACS-purified GC B cells, the purities of MACS-isolated untouched GC B cells from immunized mice are relatively low (Ramezani-Rad and Rickert, 2021), which often necessitates further enrichment by FACS via antibody labeling of MACS-enriched GC B cells, depending on the application (Ou et al., 2021).
Built upon published MACS protocols (Cato et al., 2011; Ramezani-Rad and Rickert, 2021), we have developed a MACS protocol with innovative changes to enable isolation of GC B cells of high purity from both naive and immunized mice. More specifically, we first eliminated the need for a separation column in the existing protocols by using the Dynabeads system (Invitrogen) to increase ease and scalability. Secondly, we added DNase to the incubation buffers to prevent non-specific cell adherence to magnetic beads, making the isolation of GC B cells more reproducible. Thirdly, we maximized the effectiveness of MACS by splitting the isolation process into 3 negative selection steps that progressively deplete non-B cells and non-GC B cells at each step, including repeated selection steps against CD43 and IgD. Finally, we introduced a step of Annexin V-mediated negative selection to enhance viability of isolated GC B cells. We found that this protocol can reliably isolate a 96%–98% pure B cell population that contains ≥ 98% GC B cells from both naive and immunized mice (Figure 1). Average numbers of GC B cells isolated from 2.5 × 107 splenocytes are about 9,000 in naive mice and 15,000 in immunized mice. If higher yield is desired, a 4–7-fold increase of GC B cell numbers can be achieved by skipping Annexin V treatment (Figure 2). In addition, it is possible to exclude debris by fast FACS isolation of unlabeled GC B cells, because following MACS purification, the remaining population in the forward and side scatter lymphocyte gate is almost exclusively GC B cells. For example, further enrichment by FACS may reduce debris made up of cell membranes, which otherwise may interfere with the analysis of cell membrane proteins and lipids in the isolated GC B cells. On the other hand, further FACS may not be required for applications such as cell culture or cell activity assay.
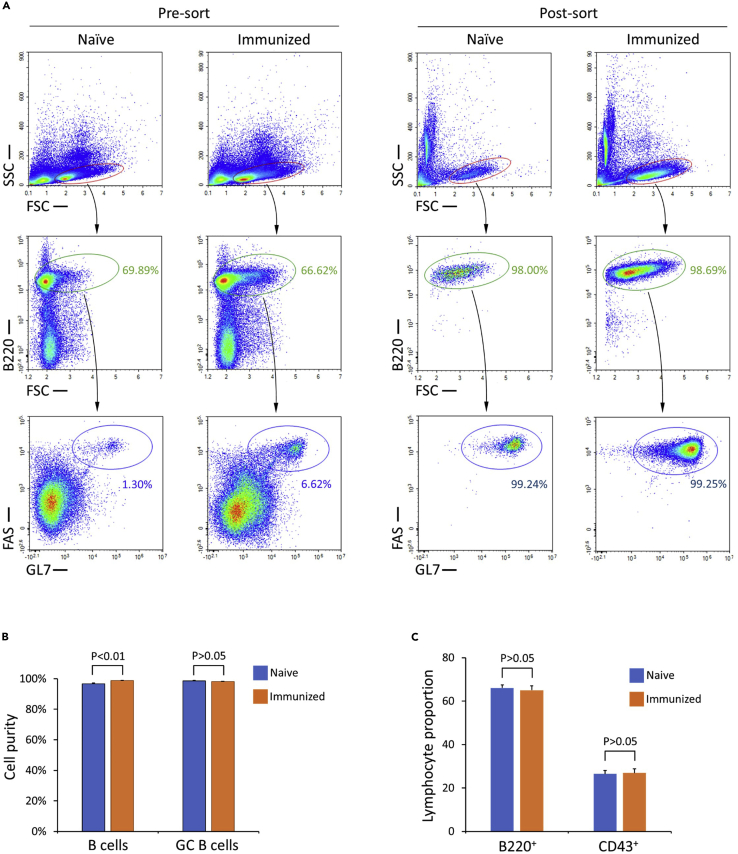
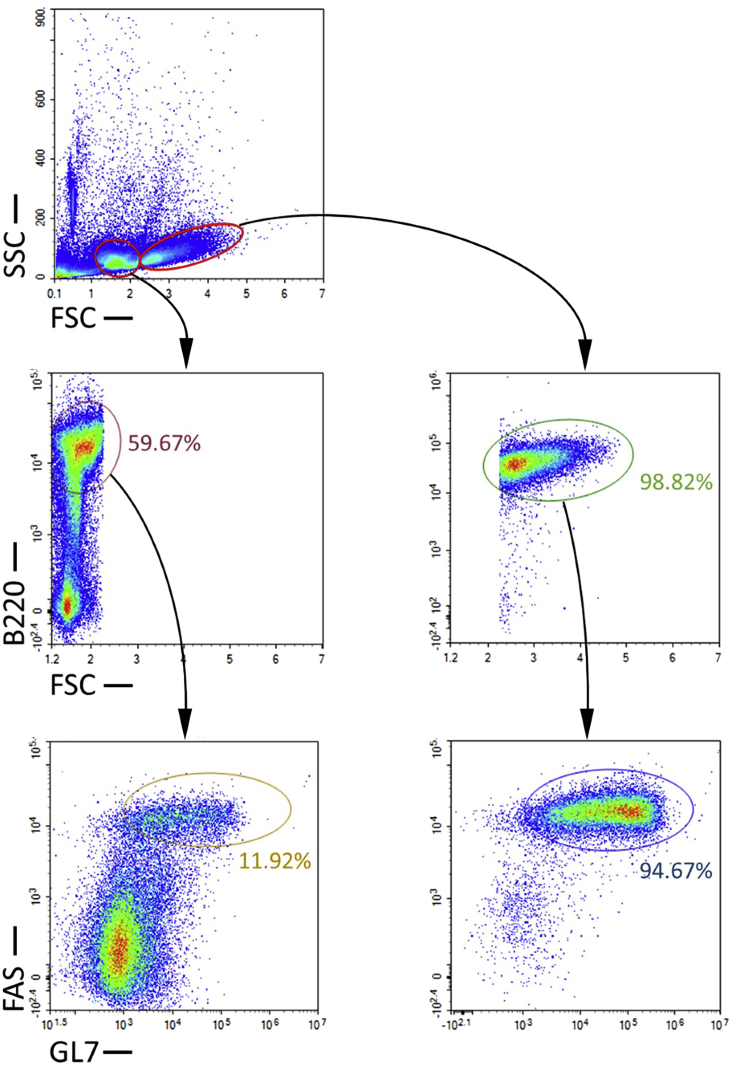
Figure 1.
Analysis of GC B cells isolated from naive and immunized mice
(A) Pre and Post Sort flow cytometry analysis of purified GC B cells from naive mice (left) and immunized mice (right). B cells are identified as B220+, and GC B cells are identified as B220+FAS+GL7+.
(B) Bar graph of compiled flow cytometry data depicting means with SEM error bars; 2-tailed T test; n = 8 biological replicates of naive and immunized mice.
(C) Bar graph showing the proportions of B220+ and CD43+ cells prior to sorting in naive and immunized mice, depicting means with SEM error bars; 2-tailed T test; n= 12 biological replicates for immunized mice; n = 13 biological replicates for naive mice.
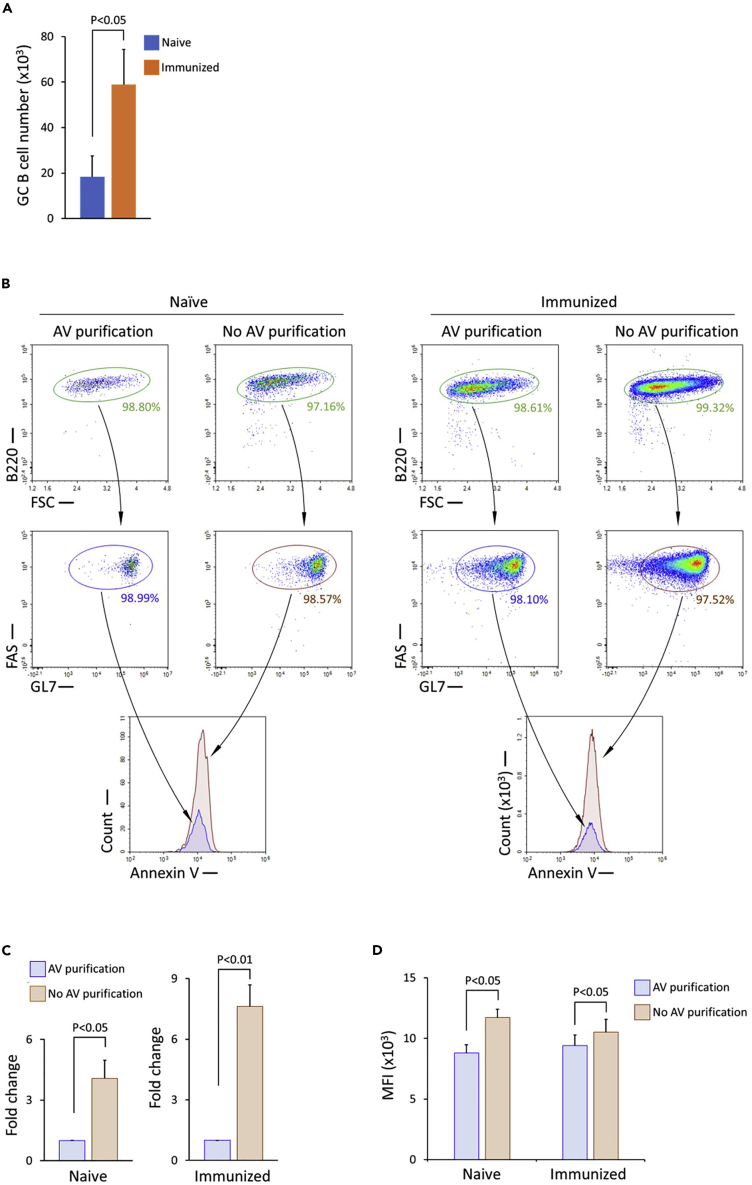
Figure 2.
Yield of GC B cells isolated from naive and immunized mice, and the effect of Annexin V-mediated negative selection on the yield
(A) Bar graph of normalized numbers of isolated GC B cells from one spleen of naive and immunized mice, depicting means with SEM error bars; 2-tailed T test; n= 8 biological replicates for immunized mice; n = 10 biological replicates for naive mice.
(B) Flow cytometry analysis of Annexin V staining intensity of purified GC B cells from naive mice (left) and immunized mice (right). The cells are separated into two halves for the treatment with and without Annexin V right before the step of Annexin V incubation to avoid the variances caused by the isolation steps prior to the Annexin V incubation. B cells are identified as B220+, and GC B cells are identified as B220+FAS+GL7+.
(C) Bar graph showing fold changes of isolated GC B cell numbers without Annexin V-mediated negative selection in naive and immunized mice, depicting means with SEM error bars; 2-tailed T test; n = 12 biological replicates for immunized mice; n = 4 biological replicates for naive mice.
(D) Bar graph of median fluorescence intensity (MFI) of Annexin V staining of GC B cells isolated from naive and immunized mice with and without Annexin V-mediated negative selection, depicting means with SEM error bars; 1-tailed T test; n = 10 biological replicates for immunized mice; n = 5 biological replicates for naive mice.
Institutional permissions
All animal experiments in this protocol were approved by the Institutional Animal Care and Use Committee of SUNY Downstate Health Sciences University. Please note that an institutional approval of animal use must be acquired before starting to use this protocol.
GC-induction
This step details the stimulation of GC formation in mice before in preparation for isolation of GC B cells.
Note: This step applies only to immunized mice. If using this protocol for unchallenged (naive) mice, proceed to the next step and harvest spleens from mice on the day of the experiment.
-
1.
Preparation and intraperitoneal injection of SRBCs have been performed as described in literature (Cato et al., 2011; Ramezani-Rad and Rickert, 2021).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotin Rat Anti-Mouse CD43 (S7) (1:50 dilution) | BD Pharmingen | Cat# 553269 RRID: AB_2255226 |
| Anti-Mouse IgD (11-26c) Biotin (1:25 and 1:50 dilution) | Thermo Fisher Scientific | Cat# 13-5993-82, RRID: AB_466860 |
| Anti-Mouse CD38 (90) Biotin (1:50 dilution) | Thermo Fisher Scientific | Cat# 13-0381-82 RRID: AB_466428 |
| Anti-Mouse CD11c (N418) Biotin (1:50 dilution) | Thermo Fisher Scientific | Cat# 13-0114-82 RRID: AB_466363 |
| Anti-Mouse TER119 (TER-119) Biotin (1:60 dilution) | Thermo Fisher Scientific | Cat# 13-5921-82 RRID: AB_466797 |
| Anti-Mouse Ly-6G/Ly-6C (Gr-1) (RB6-8C5) Biotin (1:60 dilution) | Thermo Fisher Scientific | Cat#: 13-5931-82 RRID: AB_466800 |
| Anti-Hu/Mo GL7 (GL-7) AF488 (1:100 dilution) | Thermo Fisher Scientific | Cat# 53-5902-82 RRID: AB_2016717 |
| PE Hamster Anti-Mouse CD95 (Jo2) (1:100 dilution) | BD Pharmingen | Cat#: 554258 RRID: AB_395330 |
| FITC Rat Anti-Mouse T- and B- Cell Activation Antigen (GL7) (1:100 dilution) | BD Pharmingen | Cat# 562080 RRID: AB_394981 |
| PE-Cy5 Rat Anti-Mouse CD45R/B220 (RA3-6B2) (1:100 dilution) | BD Pharmingen | Cat# 553091 RRID: AB_394621 |
| Biological samples | ||
| Citrated Sheep Red Blood Cells | Colorado Serum Company | Cat# 31102 |
| Chemicals, peptides, and recombinant proteins | ||
| Biotin Annexin V (1:50 dilution) | BD Pharmingen | Cat# 556417 RRID: AB_2869070 |
| FITC Annexin V (1:100 dilution) | BD Pharmingen | Cat# BDB556420 |
| PE Annexin V (1:100 dilution) | BioLegend | Cat# 640947 |
| APC Annexin V (1:100 dilution) | BD Pharmingen | Cat# 561012 RRID: AB_2868885 |
| Propidium Iodide Staining Solution (1:100 dilution) | BD Pharmingen | Cat# 556463 RRID: AB_2869075 |
| 7-AAD Viability Staining Solution (1:100 dilution) | eBioscience | Cat# 00-6993-50 |
| Annexin V Binding Buffer, 10× concentrate | BD Pharmingen | Cat# 556454 RRID: AB_2869074 |
| Dynabeads Mouse CD43 (Untouched B cells) | Invitrogen | Cat# 11422D |
| Dynabeads Biotin Binder | Invitrogen | Cat# 11047 |
| DNase I | Roche | Cat# 10104159001 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat# BP1605-100 |
| Fetal Bovine Serum (Heat Inactivated) | Gibco | Ref# 10082-147 |
| PBS pH 7.4 (1×) (-CaCl2) (-MgCl2) | Gibco | Ref# 10010-031 |
| eBioscience 1× RBC Lysis Buffer | Invitrogen | Ref# 00-4333-57 |
| HEPES Buffer, 1 M Solution | Fisher | Cat# BP299-100 |
| Ammonium Chloride | Fisher | Cat# A661-500 |
| Potassium Bicarbonate | Fisher | Cat# P235-500 |
| Sodium Chloride | Fisher | Cat# BP358-10 |
| Magnesium Chloride | Fisher | Cat# M33-500 |
| Calcium Chloride Dihydrate | Sigma | Cat# C5080 |
| Critical commercial assays | ||
| Debris Removal Solution | Miltenyi Biotec | Cat# 130-109-398 |
| Experimental models: Organisms/strains | ||
| Mouse: 3 to 6-month-old male and female wild type C57BL/6 | In house breeding, originally purchased from JAX lab | N/A |
| Mouse: 4-6-month-old female NZBWF1 | JAX lab | Strain#: 100008 RRID: IMSR_JAX:100008 |
| Software and algorithms | ||
| NovoExpress | Agilent | N/A |
| Excel | Microsoft | N/A |
| PowerPoint | Microsoft | N/A |
| Illustration | BioRender | N/A |
| Other | ||
| Magnetic Particle Concentrator (Dynal MPC-L) | Dynal | Product# 120.21 |
| Novocyte | Agilent | N/A |
| Eppendorf Centrifuge 5417R | Eppendorf | N/A |
| Eppendorf Centrifuge 5430R | Eppendorf | N/A |
| Sorvall ST 16R Centrifuge | Thermo Scientific | N/A |
| Frosted Microscope Slides | Fisher Scientific | Cat# 12-550-343 |
| Cell Strainer 70 μm Nylon | Corning Incorporated | Ref# 352350 |
| Labquake Rotator/Shaker | Barnstead Thermolyne | Model 40011 |
Materials and equipment
10× Annexin V Stock Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES 1 M pH 7.3 | 0.1 M | 5 mL |
| NaCl | 1.4 M | 4.09 g |
| CaCl2.H2O | 25 mM | 183.75 mg |
| ddH20 | n/a | 45 mL |
| Total | 50 mL | |
Storage at –4°C, discard if solution begins to precipitate.
Alternatives: Commercial alternatives for 10× Annexin V Buffer, such as 10× Annexin V Binding Buffer, BD Bioscience, Cat#: 556454, can be used.
Note: The following quantities are for isolating GC B cells from approximate 2.5 × 107 splenocytes, which can be harvested from ½ of the spleen (≈38 mg) of a naive mouse or ¼ of the spleen (≈21 mg) of an immunized mouse (3 to 6-month-old male and female C57BL/6); adjust volumes accordingly when working with different spleen sizes. Make the buffers fresh on experiment day and keep them on ice for the duration of the experiment.
CD43 Beads Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS (-Ca, -Mg) 1× | n/a | 1,960 μL |
| BSA (20%) | 0.1% | 10 μL |
| MgCl2 (25 mM) | 0.25 mM | 20 μL |
| DNase I (10 mg/mL) | 0.05 mg/mL | 10 μL |
| Total | 2,000 μL | |
1× Annexin V Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Annexin V Stock Solution | 1× | 120 μL |
| ddH20 | n/a | 1,080 μL |
| Total | 1,200 μL | |
Annexin V Beads Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Annexin V Buffer | 1× | 90 μL |
| ddH20 | n/a | 792 μL |
| BSA (20%) | 0.1% | 4.5 μL |
| MgCl2 (25 mM) | 0.25 mM | 9 μL |
| DNase I (10 mg/mL) | 0.05 mg/mL | 4.5 μL |
| Total | 900 μL | |
Step-by-step method details
Note: This protocol is optimized for isolating GC B cells from 2.5 × 107 splenocytes (approximate ½ splenocytes of a naive mouse or ¼ splenocytes of an immunized mouse). The timing is calculated based on the simultaneous processing of two samples. To enhance cell survival, all buffers and reagents should be kept on ice, and all centrifugations and beads incubations should be carried out at 4°C.
Harvesting splenocytes and removing red blood cells
Timing: 15–20 min
This step includes spleen removal from euthanized mice, splenocyte isolation and RBC lysis.
-
1.
Harvest mouse spleens as reported (Cato et al., 2011; Ramezani-Rad and Rickert, 2021).
-
2.
Scissor the spleen into 4–5 small segments in a 60 mm culture dish containing 1 mL of PBS.
Note: Using FBS supplemented cell culture media may enhance cell survival.
-
3.
Using minimal pressure, gently disassociate splenic tissues between the frosted ends of two glass slides to release splenocytes.
CRITICAL: Gentle release of splenocytes from fragmented splenic tissues is essential for reducing cell injury and assuring the high purity and yield of isolated GC B cells.
-
4.
Make single-cell suspension by gently pipetting up and down >10 times with a 1 mL pipette.
-
5.
Filter the cells through a 70 μm cell straining and transfer approximate 2.5 × 107 splenocytes (Equivalent to ¼ of splenocytes harvested from an immunized mouse or ½ of splenocytes harvested from a naive mouse) to an Eppendorf tube.
Note: Using < 2 × 107 splenocytes may reduce the yield and purity of isolated GC B cell. However, if using ≥ 4 × 107 splenocytes, we recommend splitting the cells into halves and isolating in two 1.5 mL tubes to avoid the decrease of the yield and purity. Alternatively, the protocol may be scaled up to isolate GC B cells from up to 3 × 108 splenocytes by using 15 mL conical tubes with proportionally increased amounts of antibodies and beads. We also suggest that a new user of this protocol should not handle more than 2 samples simultaneously.
-
6.
Pellet cells by centrifugation at 300 × g for 3 min and remove supernatant.
-
7.
Resuspend the pellet in 300 μL 1× RBC Lysis Buffer (eBioscience) and incubate on ice for 60–90 s to remove erythrocytes.
CRITICAL: Incubation with RBC lysis buffer for over 3 min may increase cell death of lymphocytes.
-
8.
Stop the lysis reaction by washing the splenocytes twice in 1 mL PBS with centrifugation at 300 × g for 3 min.
At the same time, wash CD43 Dynabeads (5 min).-
a.Transfer 100 μL beads per sample into an Eppendorf tube.
-
b.Add 1 mL of PBS and resuspend.
-
c.Place the tube on the magnet for 1 min as shown in Figure 3A.
-
d.Discard the supernatant.
-
e.Repeat wash steps b–d.
-
f.Resuspend in 200 μL CD43 Beads Buffer.
-
a.
-
9.
Resuspend the pellet in 600 μL of CD43 Beads Buffer.
CRITICAL: Without addition of DNase or MgCl2 in the beads buffer, B cells may clump/have nonspecific binding to beads due to genomic DNA present from cell injury, resulting in the loss of cells.
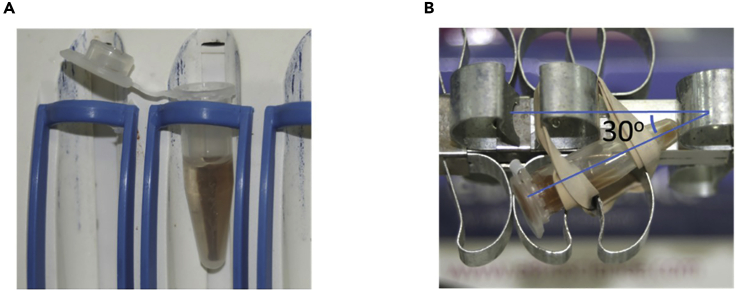
Figure 3.
Placement of the Eppendorf tube on the magnet and the rotator for cell sorting
(A) When place the Eppendorf on the magnet, ensure that the tube is closely contacted with the magnet wall. The tube can be kept open for convenience. Keep the tube on the magnet when collecting beads free supernatant.
(B) When place the Eppendorf tube on the rotator for beads incubation, the tube should be positioned with an approximate 30-degree angle to the axis of the rotator to allow smooth and constant movement of the beads in the tube.
B cell purification by negative selection of CD43+ cells
Timing: 30–35 min
This step details the enrichment of the B cell population by removal of CD43+ cells.
-
10.
Add 160 μL of pre-washed CD43 beads to the cell suspension and gently mix.
-
11.
Place the tube on the rotator as shown in Figure 3B, rotate and tilt the tube at 4°C for 20 min.
-
12.
Remove the tube and resuspend the bead-bound cells by pipetting 10 times to limit trapping of unbound B cells.
-
13.
Place the tube on the magnet for 2 min as shown in Figure 3A.
-
14.
Transfer the supernatant containing the unbound B cells to a new tube.
-
15.
Pellet cells by centrifugation at 300 × g for 3 min and resuspend in 50 μL CD43 Beads Buffer containing 3% FBS.
1st round GC B cell purification by negative selection of IgD+ cells and CD43+ cells
Timing: 75–80 min
This step details the first round of enrichment of the GC B cell population by removal of IgD+ cells and additional removal step of CD43+ cells.
-
16.
Add 2 μL biotinylated anti-IgD antibody, mix and incubate on ice for 20 min.
At the same time, prepare Biotin Binder beads (5 min).-
a.Transfer 150 μL beads into an Eppendorf tube.
-
b.Add 1 mL of PBS and resuspend.
-
c.Place tube on magnet for 1 min.
-
d.Remove supernatant.
-
e.Repeat wash steps b–d.
-
f.Resuspend the beads in 150 μL CD43 Beads Buffer.
-
a.
-
17.
Wash cells twice in 600 μL PBS by centrifugation at 300 × g for 3 min and resuspend cells in 600 μL CD43 Beads Buffer.
-
18.
Add 150 μL pre-washed Biotin Binder beads and 40 μL pre-washed CD43 beads, gently mix.
-
19.
Place the tube on the rotator as shown in Figure 3B, rotate and tilt the tube at 4°C for 20 min.
-
20.
Remove the tube and resuspend the bead-bound cells by pipetting 10 times to limit trapping of unbound GC B cells.
-
21.
Place the tube on magnet for 2 min as shown in Figure 3A.
-
22.
Transfer the supernatant containing unbound GC B cells to a new tube.
Note: Wash steps after this point in the protocol are performed at 350 × g for 5 min to minimize cell loss, as the pellet size is now significantly reduced as shown in Figure 4.
-
23.
Pellet cells at 350 × g for 5 min and resuspend in 50 μL CD43 Beads Buffer.
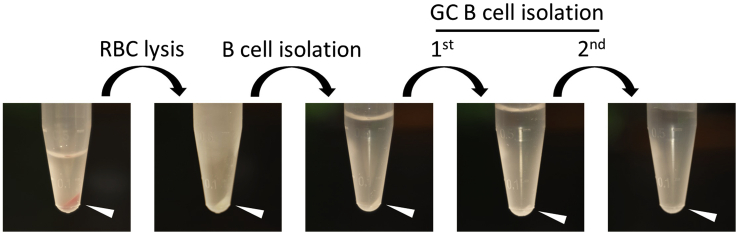
Figure 4.
Changes of cell pellet during GC B cell purification
Representative cell pellets at each step of the isolation of GC B cells from 1/4 spleen of SRBC-challenged mice. Cell pellets are indicated by white arrows. Please note that when purifying GC B cells from 1/2 spleen of a naive mouse, cell pellets may become invisible at the stages of GC B cell isolation.
2nd round GC B cell purification by negative selection of IgD+, CD11c+, CD38+ cells
Timing: 90–100 min
This step details the second round of GC B cell purification by negative selection of IgD+, CD11c+, CD38+, and Annexin V+ cells.
-
24.
Add 1 μL each of biotinylated IgD, CD11c, and CD38 antibodies, mix and incubate on ice for 15–20 min.
Optional: 0.8 μL of Ter119 and Gr1 antibodies may be added at this step to remove additional non-B cells and enhance purity further.
Note: Skipping the following steps 25 and 26 of Annexin V incubation and proceed to step 27 may dramatically increase yield of isolated GC B cells without decreasing the cell purity but may increase Annexin V+ GC B cells (Figure 2). This approach can be used if a higher yield is desired, and when the cells undergoing apoptosis do not significantly affect the downstream experiments.
-
25.
Wash with 600 μL PBS by centrifugation at 350 × g for 5 min and resuspend in 50 μL Annexin V Beads Buffer.
CRITICAL: All following steps should be conducted in Annexin V Beads Buffer or Annexin V Buffer to ensure optimal binding of biotinylated Annexin V with cells undergoing apoptosis.
-
26.Add 1 μL biotinylated Annexin V, mix and incubate on ice for 15 min.
 CRITICAL: Incubation with biotinylated Annexin V for an extended period (beyond 15 min) may result in a lower yield of GC B cells.Note: If skipping the Annexin V incubation, use CD43 Beads Buffer and PBS to replace Annexin V Beads Buffer and Annexin V Buffer respectively in the following steps.At the same time, prepare Biotin Binder beads (5 min):
CRITICAL: Incubation with biotinylated Annexin V for an extended period (beyond 15 min) may result in a lower yield of GC B cells.Note: If skipping the Annexin V incubation, use CD43 Beads Buffer and PBS to replace Annexin V Beads Buffer and Annexin V Buffer respectively in the following steps.At the same time, prepare Biotin Binder beads (5 min):-
a.Transfer 100 μL beads into an Eppendorf tube.
-
b.Add 1 mL of PBS and resuspend.
-
c.Place the tube on magnet for 1 min.
-
d.Remove supernatant.
-
e.Repeat wash steps b–d.
-
f.Resuspend in 100 μL Annexin V Beads Buffer (Use CD43 Beads Buffer instead if skipping Annexin V incubation).
-
a.
-
27.
Wash cells twice with 600 μL 1× Annexin V Buffer (Use PBS instead if skipping Annexin V incubation) by centrifugation at 350 × g for 5 min and resuspend in 600 μL Annexin V Beads Buffer (Use CD43 Beads Buffer instead if skipping Annexin V incubation).
-
28.
Add 100 μL pre-washed Biotin Binder beads, gently mix.
-
29.
Place the tube on the rotator, rotate and tilt the tube at 4°C for 20 min.
-
30.
Remove the tube and resuspend the bead-bound cells by pipetting 10 times to limit trapping of unbound GC B cells.
-
31.
Place the tube on magnet for 2 min and transfer unbound GC B cells to a new tube.
Note: Purified GC B cells are now ready for downstream applications including flow cytometry.
Expected outcomes
As shown in Figure 1, this protocol can yield highly enriched, viable and untouched GC B cells from both naive and immunized mice. More than 96% cells isolated from naive mice are B cells, in which ≥ 98% are untouched GC B cells. Similarly, more than 98% cells isolated from immunized mice are B cells, in which ≥ 98% are untouched GC B cells. Therefore, the purities of untouched GC B cells isolated by this protocol are comparable to the purities of FACS-isolated antibody-labeled GC B cells. Please note that the gating sequence used in the flow cytometry analysis here parallels the steps of GC B cell isolation and is different from that used in the published protocol (Cato et al., 2011; Ramezani-Rad and Rickert, 2021). However, this does not affect the results of purity assay (Figure S1). Interestingly, although the proportions of GC B cells in total B cells isolated from naive and immunized mice are almost identical, we noticed that the purities of B cells isolated from naive mice are slightly lower than that from immunized mice with a statistical significance (p = 0.003) (Figure 1B), despite that the proportions of B220+ B cells and CD43+ non-B cells in the splenocytes appear to be comparable between naive mice and immunized mice (Figure 1C).
As shown in Figure 2A, expected average numbers of GC B cells isolated from 2.5 × 107 splenocytes are about 9,000 in naive mice and 15,000 in immunized mice. As described in the step-by-step methods, yield can be increased 4-fold in naive mice and 7-fold in immunized mice by omission of the Annexin V incubation step (Figures 2B and 2C). While omission of this step will not affect cell purities, it can result in increased staining intensity of Annexin V in the isolated GC B cell sample, as shown in Figures 2B and 2D. However, even without the negative selection by Annexin V incubation, non-viable cells (PI+ or 7-AAD+) are nearly absent in isolated GC B cells (Figure S2). This approach should be considered when the effects of GC B cells undergoing apoptosis on the downstream experiments are negligible or can be excluded by designed control samples.
In addition to C57BL/6 mice, this protocol can be used to isolate GC B cells from other strains. As shown in Figure S2, B cells and GC B cells isolated from NZBWF1 mice without SRBC challenge have similar purities to those isolated from C57BL/6 mice (Figure 1).
Quantification and statistical analysis
Statistical details of experiments can be found in the figure legend. Data were analyzed using Excel. One- or Two-tailed T test was used to compare the two groups. Results were presented as mean + SEM and statistical significance was accepted at p < 0.05.
Limitations
Due to the nature of negative selection, the enrichment process itself cannot remove the cell debris generated during the procedure. If cell debris could affect downstream molecular and biochemical assays, such as those analyzing cell membranes, we recommend excluding the debris from untouched GC B cells by FACS, but without antibody labeling. This is feasible because following this MACS purification, the dominant cell population resolved by forward and side scatter is composed of nearly 100% GC B cells (Figure 1A), which can be cleanly gated for FACS isolation from debris with different scatter properties. Alternatively, density gradient centrifugation, such as the Debris Removal Solution (Miltenyi Biotec), can be used to partially remove cell debris, but may cause some loss of yield and death of purified GC B cells.
In addition, this protocol uses three rounds of negative selection to progressively and repeatedly remove different types of unwanted cells, which takes more time to complete compared to the existing MACS protocols (Cato et al., 2011; Ramezani-Rad and Rickert, 2021). We believe that progressive and repeated depletions are essential for increasing the efficiency of GC B cell purification in this protocol. Progressive depletion can increase bead-to-cell ratio at the following depletion steps, which can more effectively remove unwanted cells. Repeated depletion is necessary for isolating cells of higher purities. This is because once the maximum depletion efficiency is reached with a certain concentration of Dynabeads, it cannot be further increased to remove residual unwanted cells by using more beads (Dyer et al., 1998). However, in contrast to these existing protocols that use column-based MACS separation, the Dynabeads system used in this protocol does not require column purification, which makes it convenient and straightforward to scale up all reagents and volumes when working with a whole spleen or multiple mouse spleens. For example, Eppendorf tubes can be replaced by 15 mL conical tubes based on the size of samples being used, a method that has been used by members of our research group. In addition, MACS allows for the processing of multiple samples simultaneously. Therefore, this protocol can be easily adapted for the efficient isolation of large numbers of untouched GC B cells from multiple samples.
Finally, despite the effectiveness of this protocol for GC B cell isolation from naive mice, low GC B cell abundance in naive mice may affect the purity and recovery. For GC B cell isolation from mice with less than 0.5% of GC B cells in total splenic B cells, it may be necessary to scale up the sample size, pool cells from the same or different mice following the first round of GC B cell purification, and/or skip the step of Annexin V removal.
Troubleshooting
Problem 1
Low yield of GC B cells. This can be due to very low proportions of GC B cells in the starting splenocytes, harsh dissociation of splenic tissues and RBC lysis, DNA contamination, the trap of unbound B cells and GC B cells in the bead aggregates when using the magnet to sort cells, and failure to collect the cells attached to the inner wall of the Eppendorf tube (Figure 5).
Figure 5.
An example of cells attached on the tube wall
Cells on the wall are indicated by yellow arrows. Please note that the cells attached on the wall become invisible at the stages of GC B cell isolation. Always collect cells attached on the tube wall by gently washing even they are invisible.
Potential solution
Skip Annexin V negative selection step (steps 25 and 26). Scale up the starting splenocytes, reagents and volumes (step 5). Keep all buffers and reagents cold. Always process the samples on ice or at 4°C. Cut the spleen into 4–5 pieces before gently dissociating splenic tissues (steps 2 and 3). Gently resuspend splenocytes to ensure single-cell suspension (step 4). Do not incubate with RBC lysis buffer longer than 3 min (step 7). Add sufficient DNase and MgCl2 to digest genomic DNA released from injured cells. Pipet up and down at least ten times following the beads incubation and before sorting with the magnet. This may prevent unbound GC B cells from being trapped in the fraction of bead aggregates (steps 12, 20, and 30). Gently wash the inner wall of the tube after centrifugation to collect the cells attached to the wall (steps 15, 23, and 25).
Problem 2
High contamination of B220- or B220low cells in isolated B cell population as shown in Figure 6. This can be due to the insufficient removal of CD43+ splenocytes because the number of splenocytes exceeded bead capacity.
Figure 6.
An example of insufficient depletion of non-B cells and non-GC B cells
In flow cytometry analysis, B cells are identified as B220+, and GC B cell are identified as B220+FAS+GL7+.
Potential solution
Sufficient RBC lysis can avoid the interference of MACS isolation by RBCs (step 7). Deplete CD43+ cells twice by repeatedly using CD43 beads or by CD43 beads and CD43 antibodies (steps 10 and 18). Use CD38, CD11c, Ter119, and Gr-1 antibodies to further remove non-B cells in 2nd round GC B cell isolation (step 24).
Problem 3
High contamination of non-GC B cells in isolated GC B cells as shown in Figure 6. This can result from inadequate removal of IgD+ and IgDlow B cells, plasmablasts, and plasma cells.
Potential solution
Ensure to remove IgD+ B cells after the depletion of non-B cells (step 16). Reinforce the removal of IgD+ B cells by repeatedly using IgD antibody in 2nd round GC B cell isolation (step 24). CD38 and CD11C antibodies may help deplete IgDlow B cells, plasmablasts, and other non-GC B cells (step 24).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chongmin Huan (chongmin.huan@downstate.edu).
Materials availability
This study did not generate new reagents.
Acknowledgments
We thank A. Schwartzman, L. Dresner, and the Department of Surgery and Cell Biology (SUNY Downstate) for the support; J.A. Edwards for the discussion; and C. Mueller for technical support. This study was supported by DoD Lupus Research Program (Concept Award, log no. LR190106) to C.H. Institutional permissions: All animal experiments in this protocol were approved by the Institutional Animal Care and Use Committee of SUNY Downstate Health Sciences University. Please note that an institutional approval of animal use must be acquired before starting to use this protocol.
Author contributions
C.H. conceptualized research with input from S.A.D.; C.H. designed the experiments in conjunction with S.A.D., S.C., and P.O.; S.A.D. and C.H. implemented and conducted most of the experimental work in conjunction with S.C. and P.O.; S.A.D. analyzed the data with input from C.H.; S.A.D., C.H., and C.A.J.R. wrote the manuscript with input from S.C. and P.O.; and C.H. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101388.
Supplemental information
Data and code availability
This study did not generate or analyze any datasets. No code is available.
References
- Cato M.H., Yau I.W., Rickert R.C. Magnetic-based purification of untouched mouse germinal center B cells for ex vivo manipulation and biochemical analysis. Nat. Protoc. 2011;6:953–960. doi: 10.1038/nprot.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P.A., Brown P., Edward R. In: Cell Separation: A Practical Approach. Fisher D., Francis G.E., Rickwood D., editors. OUP Oxford; 1998. Immunomethods: magnetic, column, and panning techniques; pp. 191–212. [Google Scholar]
- Goillot E., Raingeaud J., Ranger A., Tepper R.I., Davis R.J., Harlow E., Sanchez I. Mitogen-activated protein kinase-mediated Fas apoptotic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3302–3307. doi: 10.1073/pnas.94.7.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou P., Stanek A., Huan Z., Roman C.A.J., Huan C. SMS2 deficiency impairs PKCδ-regulated B cell tolerance in the germinal center. Cell Rep. 2021;36:109624. doi: 10.1016/j.celrep.2021.109624. [DOI] [PubMed] [Google Scholar]
- Ramezani-Rad P., Rickert R.C. Quick and easy purification of murine untouched naive B cells or germinal center B cells by MACS. Star Protoc. 2021;2:100369. doi: 10.1016/j.xpro.2021.100369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J.F., Berkova Z., Mathur R., Zhu H., Braun F.K., Tao R.H., Sabichi A.L., Ao X., Maeng H., Samaniego F. Nucleolin inhibits Fas ligand binding and suppresses Fas-mediated apoptosis in vivo via a surface nucleolin-Fas complex. Blood. 2013;121:4729–4739. doi: 10.1182/blood-2012-12-471094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze any datasets. No code is available.