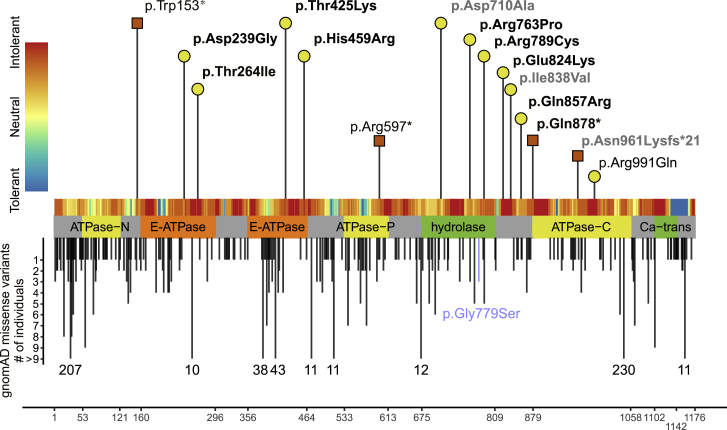
Figure 2.
Variants in ATP2B1
Location of missense and loss-of-function variant in ATP2B1 with respect to the domain structure of ATP2B1 (GenBank: NM_001001323.2). The x axis represents the corresponding amino acid position of ATP2B1. Variants reported in this study are labeled with the corresponding p-code and are indicated by a yellow circle (missense) or a red square (loss of function). Confirmed de novo variants are indicated in bold. Deciphering Developmental Disorders study variants with a lacking detailed phenotypic description are indicated in gray (see also Table S1 and supplemental methods). Missense variants in gnomAD with allele count are shown below the protein scheme. The gnomAD variant p.Gly779Ser, which was used as negative control for the Ca2+ imaging experiments, is blue color-coded. The tolerance landscape (MetaDome18) is shown color-coded above the protein scheme. Abbreviations: ATPase-N, cation transporter/ATPase, N terminus; ATPase-P, cation transport ATPase (P-type); ATPase-C, cation transporter/ATPase, C terminus; E-ATPase, E1-E2 ATPase; hydrolase, haloacid dehalogenase-like hydrolase; Ca-trans, plasma membrane calcium transporter ATPase C-terminal.