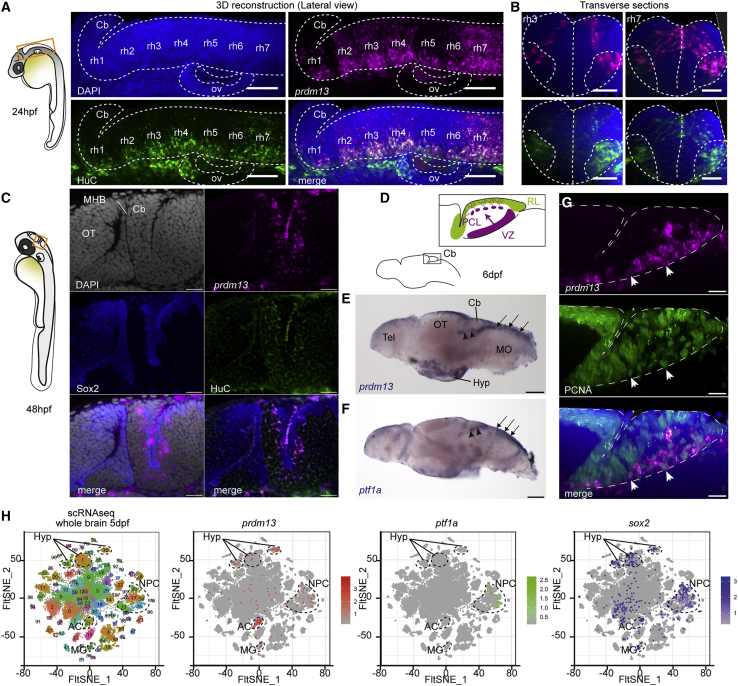
Figure 4.
prdm13 expression during zebrafish hindbrain development
(A) Lateral view of a 3D reconstruction of the zebrafish hindbrain at 24 hpf showing the expression of prdm13 (ISH, magenta) with a whole-mount immunostaining for the neuronal marker HuC (green). Cell nuclei are counterstained with DAPI (blue). A dotted line contours the hindbrain region. Cb, cerebellum; rh1–7, rhombomere 1–7; ov, otic vesicle. Scale bar: 50 μm.
(B) Transverse section through the 3D reconstruction shown in (A) at the level of rhombomere 3 (left panel) and rhombomere 7 (right panel). Scale bar: 50 μm.
(C) Optical z-plane showing prdm13 expression (magenta) together with an immunostaining for Sox2 (blue) and HuC (green) in the cerebellum at 48 hpf. Cell nuclei are counterstained with DAPI (gray). At this stage, prdm13 starts to be expressed in the cerebellar primordium, corresponding to the dorsal part of the first rhombomere, adjacent to the midbrain-hindbrain boundary (MHB). OT, optic tectum. Scale bar: 20 μm.
(D) Schematic representation of the larval zebrafish brain at 6 days post fertilization (dpf), with a magnification on the larval cerebellum. Like in mammals, the zebrafish cerebellum develops from two distinct neurogenic progenitor pools: the rhombic lip (RL, green) and the ventricular zone (VZ, purple). The VZ gives rise to all the GABAergic neuronal populations, including Purkinje neurons, which migrate and align to form the Purkinje cell layer (PCL).
(E and F) Whole-mount ISH on zebrafish larval brains at 6 dpf showing the expression of prdm13 (E) and ptf1a (F) in blue. Note the expression of prdm13 and ptf1a in the cerebellar VZ (black arrowheads) and dorsal hindbrain (black arrows). Cb, cerebellum; OT, optic tectum; Tel, telencephalon; Hyp, hypothalamus; MO: medulla oblongata. Scale bar: 100 μm.
(G) Lateral view of a 3D reconstruction of the zebrafish cerebellum at 6 dpf showing the expression of prdm13 (ISH, magenta) with a whole-mount immunostaining for the proliferation marker PCNA (green). Cell nuclei are counterstained with DAPI (blue). Dotted lines outline the cerebellar primordium. prdm13 transcripts are detected in proliferating progenitors of the VZ (white arrowheads). Scale bar: 10 μm.
(H) t-distributed stochastic neighbor embedding (t-SNE) projection plots of distinct single-cell populations obtained from the zebrafish brain at 5 dpf. The zebrafish single-cell RNA-seq (scRNA-seq) dataset was retrieved from the Gene Expression Omnibus (GEO) public database under accession GEO: GSE158142.24 In the first panel, cells are color-coded according to cluster annotations from the original publication.24 In the three other panels, cells are colored in graded intensities, reflecting the expression levels of prdm13, ptf1a, and sox2. Clusters in which prdm13 expression is enriched are circled with dotted lines. prdm13 expression is enriched in clusters 13, 25, 30, and 38, corresponding to neural progenitors (NPCs) that are also enriched for ptf1a and sox2. Expression of prdm13 is also detected in clusters 1, 34, and 35, which are annotated as hypothalamic clusters (Hyp); 21, which corresponds to amacrine cells (ACs); and 45, which correspond to Müller glia (MG).