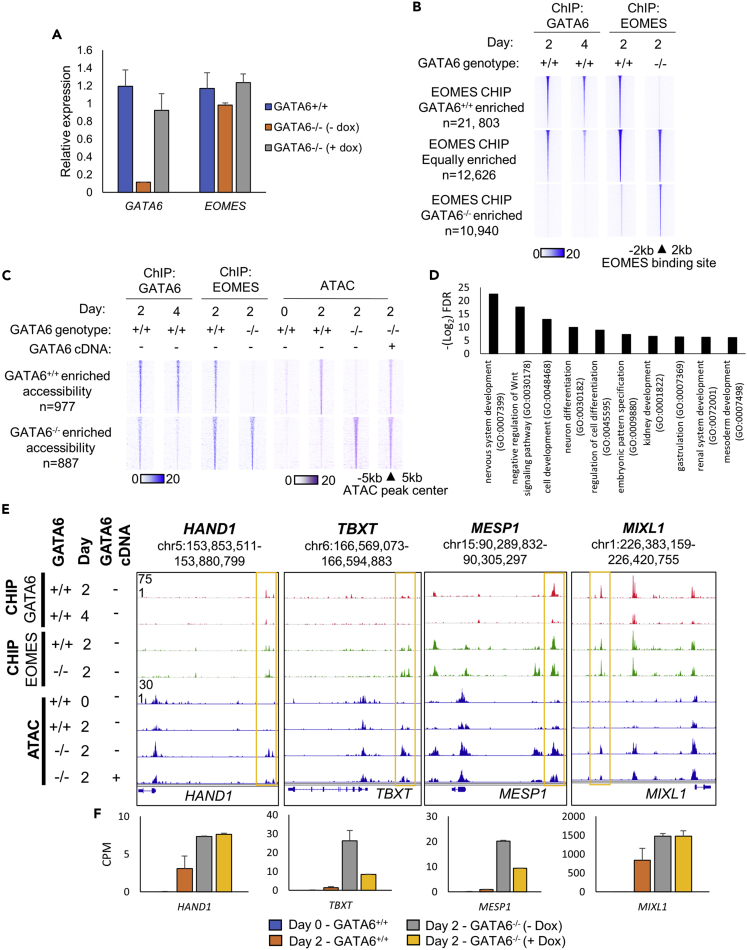
Figure 4.
EOMES occupies regions of increased chromatin accessibility in GATA6−/− cells
(A–F) RT-qPCR analysis of GATA6 and EOMES mRNA levels at day 2 of differentiation in GATA6+/+ and GATA6−/− cells ± doxycycline. Values normalized to the housekeeping mRNA RPL13a and shown relative to GATA6+/+ cells, n = 3 mean ± SD; (B) Heatmap intensity of GATA6 and EOMES ChIP-seq signal at subsets of EOMES binding that are GATA6-dependent and GATA6-independent; (C) Heatmaps depicting EOMES and GATA6 ChIP- and ATAC-seq signal intensity in GATA6+/+ and GATA6−/− cells at GATA6 occupied regions of differentially accessible chromatin identified between GATA6+/+ and GATA6−/− cells ± doxycycline during early endoderm formation; (D) GO biological process analysis. Selected significantly-enriched developmental pathways related to the genes that neighbor regions of chromatin uniquely accessible in GATA6−/− cells; (E) Genome viewer representation of the HAND1, TBXT, MESP1, and MIXL1 genomic loci aligned with GATA6 and EOMES ChIP-seq and ATAC-seq signal during early endoderm formation in GATA6+/+ and GATA6−/− cells; (F) mRNA expression of the genes that neighbor the selected examples of differentially accessible regions of chromatin, RNA-seq data displayed as counts per million (CPM), n = 2 mean ± SD.