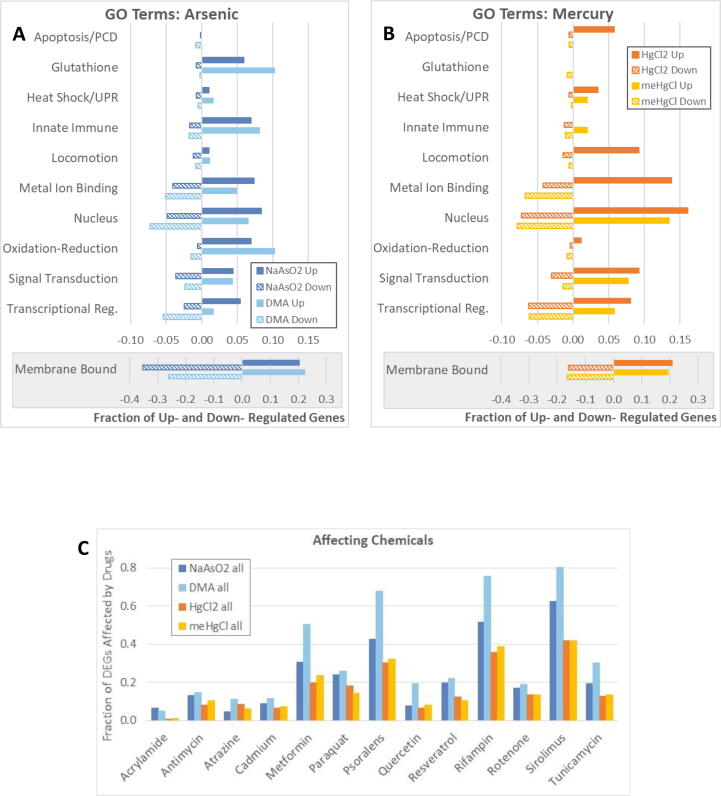
Fig. 3.
Gene Ontology (GO) terms, and genes also affected by other chemicals. WormBase’s GO terms and descriptions relating to specific functions were used to assign into categories all differentially expressed genes (DEGs) for which there was expression, function, and/or homology information in WormBase. (A-B) The number of DEGs varied among conditions, therefore bars represent the fraction of all DEGs that were in each category. Many DEGs fell into multiple categories and were therefore counted more than once. Note that the fraction of DEGs with the GO term “integral component of membrane” (Membrane Bound) are shown with a different scale. (C) The fraction of DEGs (y-axis) also altered by exposure to listed drugs (x-axis) is graphed. WormBase’s expression cluster summary only indicates that a gene’s expression is affected by a chemical, but not in which direction, therefore the fraction of DEGs also affected by various chemicals is presented for up- and downregulated DEGs together. Abbreviations: Upregulated (Up), Downregulated (Down), Programmed Cell Death (PCD), Unfolded Protein Response (UPR), Transcriptional Regulation plus Histone Modification (Transcriptional Reg.).