Summary
Background
Rheumatoid arthritis (RA) is a chronic, immune-mediated inflammatory disease of the joints that has been associated with variation in the peripheral blood methylome. In this study, we aim to identify epigenetic variation that is associated with the response to tumor necrosis factor inhibitor (TNFi) therapy.
Methods
Peripheral blood genome-wide DNA methylation profiles were analyzed in a discovery cohort of 62 RA patients at baseline and at week 12 of TNFi therapy. DNA methylation of individual CpG sites and enrichment of biological pathways were evaluated for their association with drug response. Using a novel cell deconvolution approach, altered DNA methylation associated with TNFi response was also tested in the six main immune cell types in blood. Validation of the results was performed in an independent longitudinal cohort of 60 RA patients.
Findings
Treatment with TNFi was associated with significant longitudinal peripheral blood methylation changes in biological pathways related to RA (FDR<0.05). 139 biological functions were modified by therapy, with methylation levels changing systematically towards a signature similar to that of healthy controls. Differences in the methylation profile of T cell activation and differentiation, GTPase-mediated signaling, and actin filament organization pathways were associated with the clinical response to therapy. Cell type deconvolution analysis identified CpG sites in CD4+T, NK, neutrophils and monocytes that were significantly associated with the response to TNFi.
Interpretation
Our results show that treatment with TNFi restores homeostatic blood methylation in RA. The clinical response to TNFi is associated to methylation variation in specific biological pathways, and it involves cells from both the innate and adaptive immune systems.
Funding
The Instituto de Salud Carlos III.
Keywords: Rheumatoid arthritis, TNF inhibitors, Treatment response, Epigenetics, DNA methylation
Research in context.
Evidence before this study
The efficacy of TNF inhibitors (TNFi) in rheumatoid arthritis patients is heterogeneous, ranging from clinical remission to a lack of improvement in disease activity. Understanding the biological basis for these differences in response has been elusive. There is clear evidence that peripheral blood of RA patients has a different methylation profile from that of healthy individuals. Previous studies have suggested that methylation variation in peripheral blood could also be associated with clinically relevant features, including response to therapy.
Added value of this study
In this study, we unprecedently analyzed the peripheral blood methylome of RA patients across time and we observed that TNFi therapy systematically reverts blood epigenetic variation of the disease towards that of a healthy state, and that methylation in specific biological pathways before starting therapy is associated with patient response. To our knowledge, it is also the first epigenome-wide association study (EWAS) for TNFi response to validate the results in an independent patient cohort. Using a cell type deconvolution approach, we have shown that the response to TNFi depends on the methylation profile of different cell types from the adaptive and innate immune system, with monocytes at the apex.
Implications of all the available evidence
The results from this study delineate biological pathways and immune cell types that are relevant for the efficacy of TNF inhibition therapy in RA. These results leverage future studies aimed at building biomarkers for patient stratification as well as strategies to identify efficacious therapies for non-responders to TNFi.
Alt-text: Unlabelled box
Introduction
Rheumatoid arthritis (RA) is an immune-mediated inflammatory disease and the most prevalent form of autoimmune arthritis.1 With the advent of biological disease-modifying anti-rheumatic drugs (bDMARDs) the prognosis of many patients has changed, and disease remission is now considered an achievable clinical objective.2 Despite this medical success, there is still a large fraction of patients (∼40%) who do not respond to bDMARDs.3 This lack of efficacy has many negative downstream consequences, including disease progression, increased disease risks by unnecessary immunomodulatory therapies, and a significant waste of economic resources.4 Understanding the source of this heterogeneity not only will help to direct patients to the most efficacious therapy, but also could provide insights into new therapeutic targets.
Tumor-necrosis factor inhibitors (TNFi) are currently the most commonly used bDMARD, and a first-line therapeutic approach in many health systems.5 Despite being used for over 20 years,6 little is known in regards to the biological factors that affect response to this therapy, and there is yet no biomarker available to personalize its administration. Clinical factors like sex and auto-antibody production have been weakly associated in some studies, but not replicated in many others.7 Genetic variation has been actively examined as a source of biomarkers with a similar lack of reproducibility.8 We and others have identified gene expression profiles with predictive potential of anti-TNF response in RA.9, 10, 11 However, this approach has also suffered from lack of reproducibility. While this could be due to differences in sample type (PBMC, whole blood), technology (one/two-color microarrays, RNA-seq) or study design, the intrinsic variability of gene expression could also be hindering the identification of reproducible findings. Instead, epigenetic variations constitute a much more stable biological feature, that can be even passed on through different cell generations.12 This stability might prove to be an advantageous feature to identify the biological mechanisms of patient heterogeneity associated with drug response.
The analysis of methylation variation has been used to characterize different aspects of RA pathology, including response to therapies. Genome-wide methylation analysis has shown that early in RA, synovial cells show abnormal methylation profiles which change through disease course,13,14 and these changes are associated with disease progression.15 The invasiveness of the synovial sample collection procedure together with the need of large cohorts to identify significant methylation changes are, however, a major limitation to study TNFi response in the inflamed tissue. In RA, however, strong epigenetic changes have been found in the whole blood of patients compared to controls.16 Cytokines associated with RA have been shown to shape the methylation profile of blood monocytes.17 These findings have prompted the search for epigenetic variation in blood associated with clinical outcomes of interest like disease activity, disease subtyping and response to therapy.18,19
In the present study we have analyzed the circulating methylome of RA patients to identify the presence of methylation variation associated with TNFi response. Compared to previous studies, we included crucial improvements, including the temporal analysis of methylation changes through a longitudinal design, as well as the validation of the association results in an independent patient cohort. To further characterize the methylation variation associated with TNFi in blood we have also performed cell type-specific analysis using a novel bioinformatic tool that determines DNA methylation in specific cell populations deconvoluted from bulk signatures. Using this novel statistical algorithm that incorporates cell proportions into the association analysis, we have identified the differentially methylated CpGs associated with TNFi response in the six main cell types in blood.20 Together, our results provide a clear view of the relevance of blood epigenetics for TNFi treatment stratification in RA.
Methods
Study design and patients
The patients from the discovery and validation cohorts analyzed in this study were recruited by the Immune-Mediated Inflammatory Diseases Consortium (IMIDC).21 The two longitudinal cohorts were collected at the rheumatology departments from 12 different university hospitals in Spain. In the discovery cohort, peripheral blood samples were obtained from 62 rheumatoid arthritis (RA) patients (>18 years old) starting TNFi therapy. All patients had a disease activity score for 28 joints (DAS28)22 of >3.2 at the beginning of treatment. Enrolled patients were followed up at week 12 of therapy, when a second blood sample was extracted for methylation analysis. The validation cohort consisted of 60 RA patients selected using the same features as the discovery cohort. Table 1 summarizes the characteristics of the two longitudinal cohorts. At week 12, the efficacy of TNFi was evaluated using the European League Against Rheumatism (EULAR) response criteria.22 The EULAR criteria are based in the temporal change in DAS28 and categorizes patients as Good, Moderate and None responders. For the present study, TNFi Good and Moderate patients were aggregated into a single Responder class. Week 12 was chosen as it is the time point at which a highly reliable measure of efficacy for biologic DMARDs is captured. As such, many clinical trials in RA use the response at week 12 as primary endpoint23 and furthermore, the EULAR guidelines suggest lack of efficacy of a biologic DMARD (like TNFi) at this week as an indicator to change therapy. DNA was extracted from whole blood samples collected at baseline and week 12 and stored at −80°C.
Table 1.
Clinical characteristics of the longitudinal discovery and validation cohorts of RA patients treated with TNFi.
| Discovery | Validation | |
|---|---|---|
| Total (n=62) | Total (n=59) | |
| Female, n (%) | 53 (85.8) | 50 (84.8) |
| Age, mean (SD) | 52.9(12.9) | 53.1 (14.1) |
| DAS28_Basal, mean (SD) | 5.44 (1.18) | 5.23 (1.17) |
| DAS28_w12, mean (SD) | 4.04 (1.21) | 3.14 (1.53) |
| RF, positive (%) | 46 (88.5) | 47 (78.3) |
| ACPA positive, n (%) | 49 (79) | 43 (74.1) |
| bDMARD naive, n (%) | 14 (22.6) | 19 (31) |
| Smoking, n (%) | 16 (25.8) | 15 (25.4) |
| Treatment, n (%) | ||
| Adalimumab | 5 (8) | 7 (12) |
| Certolizumab | 10 (16) | 13 (22) |
| Etanercept | 34 (55) | 31 (52) |
| Golimumab | 12 (19.3) | 9 (15) |
| Infliximab | 1 (1.6) | 0 |
SD: standard deviation; DAS28: disease activity score for 28 joints; RF: rheumatoid factor antibodies; ACPA: anti-cyclic citrullinated antibodies; bDMARD: biological disease-modifying anti-rheumatic drug.
Ethics
The study was approved by Hospital Universitari Vall d'Hebron Clinical Research Ethics Committee with reference number 20/0022. This study was conducted according to the principles of the Declaration of Helsinki. Protocols were reviewed and approved by the local institutional review board of each participating centre.
Methylome profiling
DNA (500 ng) from each sample was sodium bisulfite-treated using the EZ96 DNA methylation kit (Zymo Research) according to the manufacturer's instructions. DNA methylation levels for >850,000 CpG sites was quantified using the Illumina EPIC BeadChip. Samples were randomized within arrays to minimize confounding by batch effects. Illumina GenomeStudio software was used to extract the raw signal intensities from each probe.
Data analysis was performed in R version 4.0.4. Probes displaying a detection P-value > 0.05 in more than 5% of samples were removed. Probes containing SNPs in close proximity to the CpG site, probes in sex chromosomes, and potentially cross-reactive probes were removed using the rmSNPandCH function from the DMRcate package. Data was normalized using the preprocessNoob method available in minfi.24 The discovery and replication groups were pre-processed and normalized using the same parameters. DNA methylation levels were analyzed as beta values, which correspond to the ratio between the methylated and unmethylated probes and is approximately equal to the percentage of methylation for each site.
Computational analyses
The association of methylation variation with TNFi response in whole blood was performed using both individual CpG sites and biological pathways. In the first approach, individual CpG association analysis was conducted to identify differentially methylated positions (DMPs) at baseline and at week 12 (time of clinical response determination). For this objective, a linear regression model was fitted using the Bioconductor package limma25 adjusting for sex, cell type and age as covariates. Epigenome-wide multiple test correction was performed using Benjamini and Hochberg false-discovery rate method (FDR).
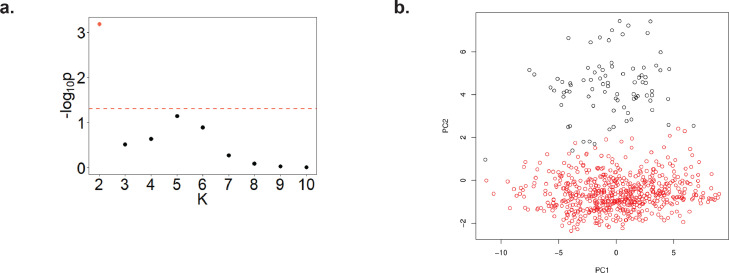
In the second analytical approach, pathway association with TNFi response was performed in two steps. In the first step, we identified the methylation variation that is associated with RA. To do this, we reanalyzed a large methylation dataset of 354 RA patients and 335 healthy controls.16 The raw data was downloaded from GEO (accession number GSE42861) and processed in the same way as the discovery and validation cohorts. Unsupervised analysis of the processed data identified an outlier subset of individuals (n=83, 12%, 53 patients and 30 controls, Figure 1) showing a distinct methylation profile. Since this group of patients was not associated with disease or other annotated clinical features, it was removed before the association analysis. Analysis of the estimated cell proportions of the main cell subsets, did show major cell frequency shifts between the outlier cluster and the main case-control cluster, particularly an increase in CD8+ T and B cells in the former group (Supplementary Figure 1). DMPs were subsequently identified using a linear regression model adjusting for sex and age. In the second step, we determined the methylation association at the pathway level. To do this, we used the gometh function implemented in the Bioconductor package missMethyl. This method has been specifically designed to test for pathway association using array methylation data. Briefly, this algorithm has been designed to overcome two main biases present in this type of data: i) the non-homogeneous distribution in the genome of the array probe sets, and ii) the annotation of CpGs to more than one gene.26,27 Following previous studies,28 the 10,000 most significantly associated CpGs were used for pathway analysis. The pathways annotated in the Gene Ontology ‘biological process’ database were selected. Pathways with less than 10 genes and more than 300 genes were filtered out. A total of 5,926 pathways were finally tested for association, and correction for multiple testing was performed using FDR adjustment.
Figure 1.
Principal component analysis (PCA) of the RA and healthy control cohort. PCA revealed a group of individuals showing an outlier methylation profile. (a). Clustering analysis using the Partitioning Around Medoids implemented in M3C software supported the presence of two groups of individuals showing significantly different methylation profile. The x-axis indicates each of the clustering partitions evaluated (k=2 to 10); k=2 (orange dot) showed a highly significant evidence for clustering (P<0.001). (b). Principal components of the RA and control cohort showing the outlier group of individuals in the second PC, and color-coded based on the M3C k=2 class assignments.
The longitudinal design of our study allowed us to interrogate the changes in DNA methylation pathway profiles induced by TNFi therapy and whether these changes drive the profiles to ones that are similar of healthy individuals. To test this, we developed an analysis method that integrates both the significance and the direction of the methylation changes. Briefly, for each pathway, we identified all the CpGs mapping to its genes, and selected those that were significantly associated with RA as well as with the longitudinal effect of TNF (nominal P<0.05). Next, we categorized the list of CpGs as hyper- and hypomethylated for the two conditions and compared them using a 2×2 contingency table. Using a Fisher's test, we determined if the methylation induced by TNFi was more similar to RA (OR>1, P<0.05) or more similar to healthy individuals (OR<1, P<0.05).
The pathway analysis method used in this study is powerful as it avoids biases that are particular to methylation data. In the significantly replicated pathways, we sought to provide an additional measure of the consistency of the methylation variation between the discovery and validation cohorts. For this objective, we developed a resampling-based test. In this method, the CpGs mapping to the genes of each pathway were first retrieved using the Illumina Infinium EPIC annotation. From the resulting list of CpG sites, we identified the number of associations that were significant (P<0.05) and in the same direction (i.e. log fold change of the same sign) in both the discovery and validation cohorts. Next, we obtained an empirical estimate of the probability of observing this number of consistent associations given the number of genes tested for each pathway. To do this, we performed a random resampling (N=1,000) of gene sets of the same number of genes as the original pathway, and calculated the number of CpGs showing significant (P<0.05) and consistent methylation changes (i.e. same direction) in the discovery and validation cohorts. The empirical P-value was then obtained by determining the fraction of resamplings with a higher consistency compared to the original observation.
In order to find differentially methylated positions for the major cell types present in blood, we applied CellDMC.20 CellDMC is a linear regression framework that integrates cell-type information to identify cell-specific methylation association. Testing for the statistical interaction between the phenotype (here response to TNFi) and the different cell proportions in the mixture, this method can reliably identify cytosines that are differentially methylated within a specific cell type. CellDMC has shown superior sensitivity and specificity compared to other approaches20 and has led to the identification of strong and reproducible cell-specific associations.29 In the present study, cell type fractions for the six most abundant cell types in blood -neutrophils, macrophages, CD4+ T cells, CD8+ T cells, B cells and NK cells- were estimated using a commonly used blood reference-based method30 and implemented in the estimateCellCounts function from the minfi package. Adjustment for multiple testing of the resulting DMPs using CellDMC was performed using Benjamini-Hochberg FDR method.
Using the cell-specific DMPs associated with TNFi response (FDR < 0.05), we conducted a motif enrichment analysis of the hypermethylated and hypomethylated DMPs associated using the PWMenrich Bioconductor package.
Role of the funding source
The present study was funded by the Instituto de Salud Carlos III, with grant number PI15/00424 (Co-funded by European Regional Development Fund/European Social Fund). The sponsor of the study had no role in the design, data collection, analysis or interpretation; in the writing of the manuscript, or in the decision to submit the paper for publication.
Results
RA patient cohorts and methylation determination
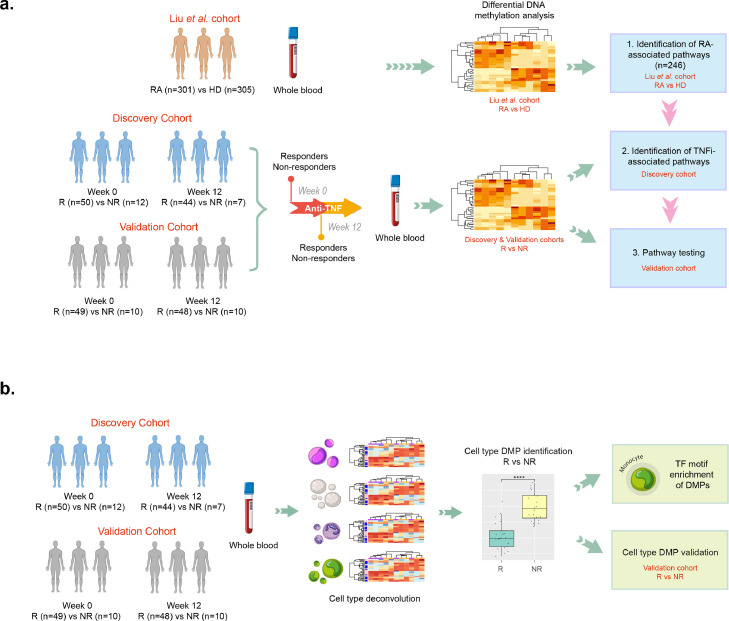
Prompted by previous association results of blood DNA methylation with the response to etanercept,31 we performed a study for the identification of epigenetic variation associated globally to TNF inhibition in a larger cohort of RA patients (n=62 patients, Table 1). Whole blood methylation was measured at the initiation of therapy (week 0) and at the determination of the clinical response to treatment 3 months later (week 12). Validation of the results was performed using a second longitudinal cohort of n=60 RA patients starting a TNFi. The two cohorts did not present significant differences related to age (P = 0.91, t-test), gender (P=0.81, Fisher's test), baseline disease activity (P=0.34, t-test), seropositivity to ACPA and rheumatoid factor (P=0.51 and P=0.39, respectively, Fisher's test), proportion of bDMARD naive patients (P=0.06, Fisher's test), or smoking (P=0.26, Fisher's test). Also, within each cohort, responder and non-responder patients did not show significant differences other than the expected disease activity at week 12 (Supplementary Tables 1 and 2). DNA from the whole blood samples was extracted in all cases and hybridized to a DNA methylation microarray that interrogates almost 850,000 CpG sites of the human genome. This is a cost-effective method allows us to interrogate one or more CpGs per gene, which is an improvement to the more commonly used array, the Infinium HumanMethylation450K BeadChip. However, one of the main limitations of this technology is that it only interrogates ∼2% of all CpGs in the human genome, and a method such as whole genome bisulfite sequencing may provide a more complete view of the global epigenome. Figure 2 summarizes the analytical design followed in the present study to identify epigenetic variation associated with the response to TNF inhibition.
Figure 2.
Schematic representation of the analytical design used to identify methylation variation associated with TNFi response. (a). Using blood methylation data from a large case-control cohort, the biological pathways associated with RA were identified (n=246). Using these disease-linked pathways, we were able to identify biological processes that are modified by TNFi treatment in longitudinal cohort of RA patients starting therapy (n=62). Also, pathways associated with the response to therapy at week 12 were identified. Using an independent patient cohort (n=59), the findings could be validated. (b). Using a novel cell-deconvolution approach in the discovery and validation cohorts, we were able to identify differentially methylated positions in multiple immune cell types associated with the response to TNFi. Monocytes showed the larger number of validated associations and we conducted TF motif enrichment to characterize the principal differentiation programs associated with response in this innate immune cell type.
Whole blood methylation association: site and pathway association
The whole blood DNA methylation analysis of the n=62 RA patients of the discovery cohort at baseline identified 10,001 CpG sites associated with TNFi response at the nominal level (P<0.01, Supplementary Table 3), but none remained significant after FDR correction. Based on previous evidence, we considered the possibility of methylation association to be specific to the type of TNF inhibitor.32 In order to retain sufficient statistical power, we split the discovery cohort between etanercept (n=34 patients) and the remaining TNFi drug types (n=28 patients). Stratification by type of TNFi did not lead to genome-wide FDR-corrected significant associations (Supplementary Tables 4 and 5). Of relevance, none of the 5 CpG sites previously associated with etanercept were replicated in the discovery cohort (Supplementary Table 6). Similarly, analysis of DNA methylation data week 12 also failed to identify significantly associated DMPs (Supplementary Table 7). Given that non-responders constitute a small proportion of treated RA patients (∼20%), it is possible that our cohort does not generate sufficient statistical power to detect subtle methylation differences following multiple testing adjustment. Hence, these results suggest that the methylation variation in blood associated with the response to TNFi is of small effect size and, consequently, more powerful strategies are needed to capture relevant epigenetic features.
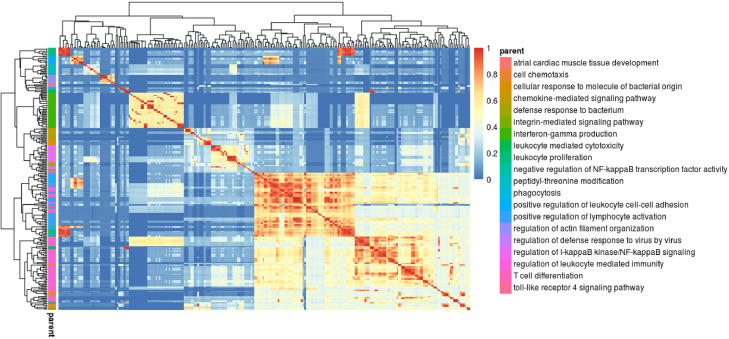
In order to increase the power to identify drug-associated methylation variation we performed a pathway analysis on biological processes associated to RA. In this approach, we assumed that the biological mechanisms that are associated with the disease (i.e. identified by comparing patients against healthy controls) may also influence the response to therapy. Previous evidence at the genetic and transcriptomic level support this possibility.33,34 Analysis of these disease-relevant biological processes at the pathway level also enables a less constrained testing of methylation variation, capturing methylation changes in genes and CpG sites other than the ones initially associated with disease. Importantly, this approach also substantially reduces the multiple testing burden (e.g. 787,087 single CpG vs 5,926 pathway tests), enabling an increase in statistical power to detect relevant associations. To identify the pathways associated with RA, we analyzed a large cohort of RA patients and healthy controls (n=301 and n=305, respectively)16 using the gometh method. A total of n=246 biological processes were found to be significantly associated with RA (FDR<0.05, Figure 3). As expected, multiple GO terms were associated to key immune-related processes including cytokine production, immune cell proliferation and differentiation.
Figure 3.
Clustering of pathways associated with RA. Comparing the methylation profile of n=301 patients and n=305 healthy individuals, a total of 246 biological processes were found to be associated with RA using gometh. In this heatmap, the associated GO terms (rows and columns) are clustered according to their similarity based on Lin's measure [38]. The terms are then hierarchically clustered using complete linkage, and the tree is cut at the desired threshold (here 0.7). For each resulting cluster, the biological process showing the most significant association with RA was chosen as the representative biological function (right legend). A total of n=20 pathway clusters were identified associated with RA.
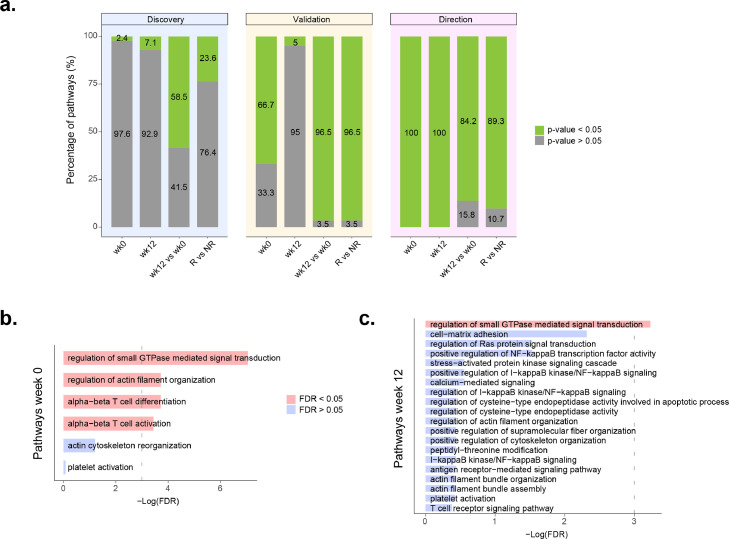
We next seek to determine the longitudinal impact of systemic TNF blocking globally, irrespective of the clinical response at the endpoint. Comparing the changes from week 0 to week 12 of TNFi treatment, we found 144 of the 246 pathways (58.5%) to be differentially methylated in the discovery cohort (FDR < 0.05, Supplementary Table 8 and Figure 4). The methylation direction test indicated that in all of the associated pathways the direction of methylation changes were towards those of healthy individuals (Figure 5 and Supplementary Table 9). Analysis of the 144 pathways in the validation cohort corroborated the association of 139 of them with TNFi therapy (96.5%). From these, resampling testing of the methylation direction analysis supported the same methylation changes in 117 pathways (84.2%, Figure 4A). Together, these results demonstrate a major impact of TNFi in the methylation profile of whole blood in active RA patients, and also show that this change is systematically in the direction of the healthy (homeostatic) state.
Figure 4.
RA pathway association with TNFi response. A. Percentage of pathways linked to RA (n=246) that were significantly associated to the discovery cohort (left bar plots). From these, the middle bar plots indicate the percentage of significantly validated pathways in the validation cohort. Finally, the right bar plots show the percentage of validated pathways that have consistent methylation changes between the two patient cohorts using the resampling-based test. wk0: pathways associated with TNFi response at week 0; wk12: pathways associated with TNFi response at week 12; wk12 vs wk0: pathways changing from week 0 to week 12 of TNFi therapy; R vs NR: pathways showing longitudinal methylation differences between responders and non-responders to TNFi therapy. B. Association results in the validation cohort of the RA pathways associated with TNFi response at week 0 in the discovery cohort. Four pathways were found to be significantly replicated (red bars, FDR < 0.05). C. Association results in the validation cohort of the RA pathways associated with TNFi response at week 12 in the discovery cohort. One pathway, regulation of small GTPase signal transduction was found to be significantly replicated (red bar, FDR < 0.05).
Figure 5.
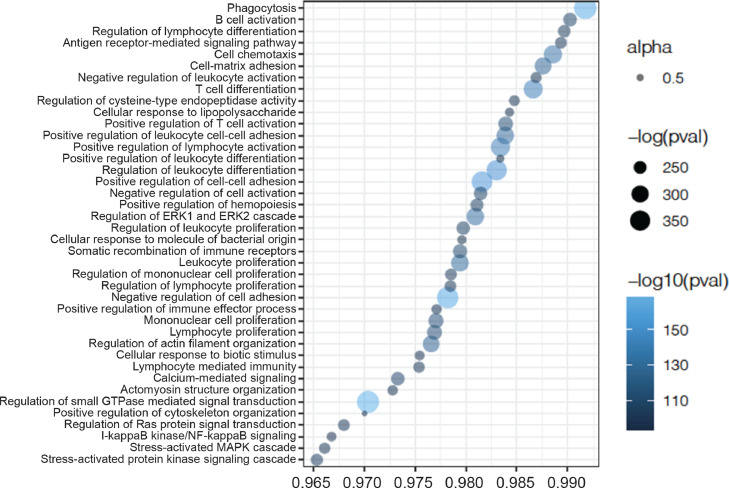
Directionality test results for pathway methylation changes induced by TNFi. Profile plot showing the percentage of CpGs having opposite methylation changes between disease development (i.e. RA vs controls) and the response to TNFi (i.e. week12 vs baseline) for the pathways linked with RA. The top 40 pathways showing the strongest significant statistical evidence are included in the plot. These results confirm the effect of blood methylation variation induced by TNFi, clearly reversing it towards that of healthy individuals.
We next tested if RA pathways are also associated with the clinical response to TNFi. In the discovery cohort we found that n=6 RA pathways were significantly different at week 0 between responders and non-responders (2.4%, FDR < 0.05, Figure 4A). Testing of these pathways in the validation cohort replicated the association of four of them with response (FDR < 0.05): “alpha-beta T cell activation”, “alpha-beta T cell differentiation”, “regulation of small GTPase mediated signal transduction” and “regulation of actin filament organization” (Figure 4B, Table 2). The resampling test confirmed that the methylation changes in all four replicated pathways were concordant between both patient cohorts (P<0.001, Figure 4A).
Table 2.
RA pathways associated with TNFi response at baseline and at week 12. Four biological pathways linked to RA were found to be differentially methylated between TNFi responders and non-responders before starting the treatment. The association was identified in the discovery patient cohort and replicated in the validation patient cohort. One of these pathways, regulation of small GTPase mediated signal transduction (GO:0051056), had a persistent differential methylation after 12 weeks of TNFi therapy. The direction of methylation changes was consistent between the discovery and validation datasets. GO: gene ontology; FDR: false-discovery rate adjusted p-value of association with TNFi response (validation cohort); emp. P-value: empirical p-value for the replication of the direction of methylation changes associated with drug response.
| Comparison | GO id | GO term | FDR | emp. P-value |
|---|---|---|---|---|
| Baseline | GO:0046631 | alpha-beta T cell activation | 0.032 | 0.029 |
| GO:0046632 | alpha-beta T cell differentiation | 0.024 | 0.002 | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | <0.001 | <0.001 | |
| GO:0110053 | regulation of actin filament organization | 0.024 | 0.003 | |
| Week 12 | GO:0051056 | regulation of small GTPase mediated signal transduction | 0.039 | <0.001 |
Analysis of differential methylation between TNFi response groups at week 12 in the discovery cohort identified n=20 significantly associated pathways (7.1%, Supplementary Table 10, Figure 4A). Testing of these pathways in the validation cohort, we replicated the association of “regulation of small GTPase mediated signal transduction” pathway after multiple test adjustment (FDR=0.039, Figure 4C). The resampling test confirmed that the methylation differences between responders and non-responders in this pathway occurred in the same direction in both patient cohorts (P<0.001).
Finally, we compared the longitudinal methylation changes between TNFi responders and non-responders. Comparing the methylation variation from week 0 to week 12 we found a total number of 58 pathways (23.6%) with a different temporal evolution according to response (FDR<0.05, Figure 4A, Supplementary Table 11). Analysis in the validation cohort replicated 56 (96.5%) of the associated temporal changes, from which 50 (89.3%) showed a statistically concordant change with the discovery cohort (P < 0.05).
Cell-specific methylation association with the response to TNFi
Following the association analysis of methylation in whole blood, we sought to test for association of methylation variation in specific blood cell types with the response to therapy. Analysis of cell-specific DMPs with CellDMC identified multiple CpG sites associated with the response to therapy at baseline in all six immune cell types analyzed. In the discovery cohort, FDR-significant DMPs were found principally in monocytes (n=2,325), followed by NK (n=1,095), CD8+T (n=940), CD4+T (n=787), neutrophils (n=253) and B cells (n=111) (Supplementary Table 12). Testing these CpGs in the patient validation cohort, 18 were replicated (P<0.05, same methylation direction) and with an epigenome-wide significance after combining the two cohorts (P<9.42E-8). Validated methylation sites included 11 CpG from monocytes, 3 from NK cells, 2 from CD4+ T cells and 2 from neutrophils (Table 3). Analysis of differential methylation at week 12 identified a larger number of DMPs associated with TNFi response, this time having a larger impact in NK (n=24,131), followed by CD4+T (n=8,706), CD8+T (n=7,049), monocytes (n=1,540), B cells (n=1,392) (Supplementary Table 13), and finally neutrophils (n=755). Association analysis of these methylation sites with TNFi response in the validation cohort also reported broader validation than week 0, with 38 CpGs replicated in NK cells, 24 in neutrophils, 21 in B cells, 19 in monocytes, 13 in CD8+T and 6 in CD4+T cells, all at epigenome-wide significance (Supplementary Table 14).
Table 3.
Cell-specific methylation sites associated with TNFi response at baseline.
| Cell type | CpG | Chr | Position | βDisc | P-valueDisc | βVal | P-valueVal | P-valueComb | Annotation |
|---|---|---|---|---|---|---|---|---|---|
| CD4+T | cg082501811 | chr3 | 158441366 | 2.14 | 1.52E-005 | 1.57 | 0.0001 | 3.85E-008 | RARRES1 |
| cg21816292 | chr8 | 24857432 | -1.46 | 7.50E-06 | -1.79 | 0.0001 | 2.31E-08 | N_Shore | |
| Mono | cg27352090 | chr2 | 218200655 | 3.37 | 5.67E-009 | 0.83 | 0.04 | 5.22E-009 | DIRC3 |
| cg03327816 | chr4 | 140879844 | 2.63 | 4.08E-006 | 2.51 | 3.21E-006 | 3.41E-010 | MAML3 | |
| cg04915579 | chr5 | 37922428 | 1.35 | 2.22E-006 | 0.7 | 1.81E-005 | 1.00E-009 | OpenSea | |
| cg19856013 | chr6 | 51342462 | 2.86 | 3.90E-009 | 0.42 | 0.012 | 1.21E-009 | OpenSea | |
| cg05056024 | chr6 | 126869507 | 2.65 | 3.03E-007 | 0.91 | 0.01 | 9.05E-008 | OpenSea | |
| cg03626668 | chr11 | 66676568 | 3.79 | 5.64E-005 | 3.3 | 4.1E-005 | 4.84E-005 | PC | |
| cg13978095 | chr12 | 14264417 | -3.21 | 1.24E-008 | -2.5 | 3.33E-009 | 1.60E-015 | OpenSea | |
| cg01104961 | chr12 | 81672471 | 2.98 | 1.14E-007 | 0.82 | 0.04 | 9.10E-008 | PPFIA2 | |
| cg00674681 | chr14 | 50517357 | 2.49 | 1.93E-006 | 1.23 | 0.002 | 7.58E-008 | OpenSea | |
| cg03719830 | chr20 | 30699752 | 2.45 | 3.16E-008 | 0.58 | 0.046 | 3.11E-008 | TM9SF4 | |
| cg22211329 | chr22 | 33504179 | 3.76 | 4.38E-006 | 1.32 | 0.0005 | 5.02E-008 | OpenSea | |
| Neu | cg209000361 | chr10 | 45938670 | 0.24 | 2.23E-006 | 0.04 | 0.0002 | 9.21E-009 | ALOX5 |
| cg268693621 | chr11 | 105947257 | -0.23 | 1.89E-006 | -0.09 | 0.001 | 3.32E-008 | KBTBD3 | |
| NK | cg05583200 | chr6 | 76109279 | 4.95 | 1.19E-008 | 4.6 | 0.006 | 1.65E-009 | FILIP1 |
| cg23224666 | chr6 | 127796287 | 3.05 | 6.40E-09 | 1.6 | 0.001 | 2.34E-10 | C6orf174 | |
| cg03957547 | chr16 | 9524433 | 6.05 | 4.90E-08 | 4.07 | 0.015 | 1.64E-08 | OpenSea | |
Using the CellDMC deconvolution approach, a total of n=18 CpGs showed a reproducible methylation association with TNFi response at baseline (P<9.42e-8). The associated CpG sites involved CD4+ T lymphocytes from the adaptive immune system and monocytes (Mono), neutrophils (Neu) and NK cells from the innate immune system. βDisc and βVal are standardized regression coefficients from the discovery and validation cohort analysis, and P-valueDisc and P-valueVal are the P-values form these analyses. P-valueComb corresponds to the P-value for the meta-analysis of the two cohorts using Fisher's method. Chr: chromosome; Annotation: biological annotation for the CpG site
Analysis of motif enrichment in monocyte TNFi response signature
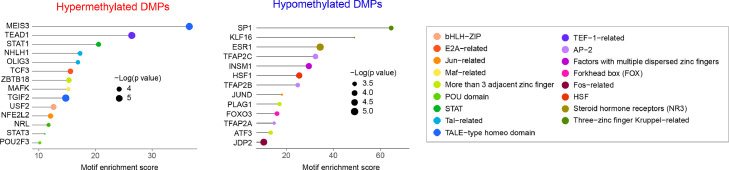
Given the observed predominant role of monocytes with TNFi response at baseline, we sought to identify the presence of enriched regulatory variation underlying this association. Regulatory motif analysis identified several transcription factors significantly associated with hyper- and hypomethylation variation in monocytes (Figure 6, Supplementary Tables 15 and 16). In this analysis, we found a significant motif enrichment for key immune signaling transcription factors STAT1 and SP1.
Figure 6.
Transcription factors associated with the response to TNFi in monocytes. Using motif-enrichment analysis on the CpG sites associated with TNFi response in monocytes, we identified multiple transcription factors significantly associated with the regulatory changes. A 500 bp region centered around the associated CpG sites in monocytes was used in the analysis. Analysis of differentially methylated positions (DMPs) was performed separately for hypermethylated and hypomethylated CpGs in responders. The motif enrichment score for each transcription factor family (TF) is depicted as a horizontal line; at the end of each line, the significance of the TF association is represented as the diameter of the circle.
Comparison to previous transcriptomic evidence on TNFi response
Recent evidence supports that treatment with infliximab, one type of TNFi, ameliorates the signatures of RA patients at the transcriptomic, proteomic and cell abundance levels.35 We evaluated if the changes at the epigenetic level mirror those at the RNA level. Comparing the epigenetic changes with this previously reported transcriptomic study, we found that from the 99 pathways modified in responders, 73 (73.7%) are also significantly altered at the RNA level (FDR < 0.05, Table 4) compared to 11 pathways modified at the RNA level (FDR < 0.05, Table 5) from the 57 differentially methylated pathways in non-responders.
Table 4.
Pathways modified in TNFi responders at the epigenetic and transcriptomic levels.
| ID | Description | P.adj |
|---|---|---|
| GO:0030217 | T cell differentiation | 9.63E-04 |
| GO:1902105 | regulation of leukocyte differentiation | 1.30E-03 |
| GO:0032943 | mononuclear cell proliferation | 1.56E-03 |
| GO:0045580 | regulation of T cell differentiation | 1.56E-03 |
| GO:0045619 | regulation of lymphocyte differentiation | 1.56E-03 |
| GO:0046651 | lymphocyte proliferation | 1.56E-03 |
| GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1.56E-03 |
| GO:0042113 | B cell activation | 1.72E-03 |
| GO:0002695 | negative regulation of leukocyte activation | 1.96E-03 |
| GO:0050866 | negative regulation of cell activation | 1.96E-03 |
| GO:0002724 | regulation of T cell cytokine production | 2.19E-03 |
| GO:0070661 | leukocyte proliferation | 2.19E-03 |
| GO:0051250 | negative regulation of lymphocyte activation | 2.32E-03 |
| GO:0046631 | alpha-beta T cell activation | 2.78E-03 |
| GO:0050852 | T cell receptor signaling pathway | 2.78E-03 |
| GO:0002369 | T cell cytokine production | 2.83E-03 |
| GO:0050851 | antigen receptor-mediated signaling pathway | 3.04E-03 |
| GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3.78E-03 |
| GO:0022409 | positive regulation of cell-cell adhesion | 3.78E-03 |
| GO:0050870 | positive regulation of T cell activation | 3.78E-03 |
| GO:0002706 | regulation of lymphocyte mediated immunity | 3.80E-03 |
| GO:0002700 | regulation of production of molecular mediator of immune response | 4.10E-03 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 4.10E-03 |
| GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 4.44E-03 |
| GO:0002699 | positive regulation of immune effector process | 5.16E-03 |
| GO:0002705 | positive regulation of leukocyte mediated immunity | 5.75E-03 |
| GO:0002708 | positive regulation of lymphocyte mediated immunity | 6.98E-03 |
| GO:0046632 | alpha-beta T cell differentiation | 6.98E-03 |
| GO:0002819 | regulation of adaptive immune response | 7.47E-03 |
| GO:0006909 | phagocytosis | 7.47E-03 |
| GO:0046578 | regulation of Ras protein signal transduction | 7.53E-03 |
| GO:0035710 | CD4-positive, alpha-beta T cell activation | 7.64E-03 |
| GO:0032729 | positive regulation of interferon-gamma production | 8.20E-03 |
| GO:0002703 | regulation of leukocyte mediated immunity | 8.75E-03 |
| GO:0002702 | positive regulation of production of molecular mediator of immune response | 9.16E-03 |
| GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1.20E-02 |
| GO:0007162 | negative regulation of cell adhesion | 1.25E-02 |
| GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1.26E-02 |
| GO:0045058 | T cell selection | 1.26E-02 |
| GO:0042093 | T-helper cell differentiation | 1.29E-02 |
| GO:0002440 | production of molecular mediator of immune response | 1.41E-02 |
| GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 1.49E-02 |
| GO:0030888 | regulation of B cell proliferation | 1.49E-02 |
| GO:0046634 | regulation of alpha-beta T cell activation | 1.53E-02 |
| GO:0002287 | alpha-beta T cell activation involved in immune response | 1.54E-02 |
| GO:0002293 | alpha-beta T cell differentiation involved in immune response | 1.54E-02 |
| GO:0002821 | positive regulation of adaptive immune response | 1.54E-02 |
| GO:0002698 | negative regulation of immune effector process | 1.69E-02 |
| GO:0046330 | positive regulation of JNK cascade | 1.77E-02 |
| GO:1903708 | positive regulation of hemopoiesis | 1.77E-02 |
| GO:0042098 | T cell proliferation | 2.17E-02 |
| GO:0046328 | regulation of JNK cascade | 2.40E-02 |
| GO:0050868 | negative regulation of T cell activation | 2.40E-02 |
| GO:1902107 | positive regulation of leukocyte differentiation | 2.61E-02 |
| GO:0051403 | stress-activated MAPK cascade | 2.72E-02 |
| GO:0032946 | positive regulation of mononuclear cell proliferation | 3.07E-02 |
| GO:0042092 | type 2 immune response | 3.07E-02 |
| GO:0050670 | regulation of lymphocyte proliferation | 3.07E-02 |
| GO:0050671 | positive regulation of lymphocyte proliferation | 3.07E-02 |
| GO:0070665 | positive regulation of leukocyte proliferation | 3.07E-02 |
| GO:0001776 | leukocyte homeostasis | 3.08E-02 |
| GO:0002449 | lymphocyte mediated immunity | 3.08E-02 |
| GO:0032944 | regulation of mononuclear cell proliferation | 3.12E-02 |
| GO:0033077 | T cell differentiation in thymus | 3.57E-02 |
| GO:0001959 | regulation of cytokine-mediated signaling pathway | 3.62E-02 |
| GO:0038094 | Fc-gamma receptor signaling pathway | 3.62E-02 |
| GO:0070663 | regulation of leukocyte proliferation | 3.62E-02 |
| GO:0031098 | stress-activated protein kinase signaling cascade | 3.72E-02 |
| GO:0007254 | JNK cascade | 3.72E-02 |
| GO:0031532 | actin cytoskeleton reorganization | 4.09E-02 |
| GO:0045582 | positive regulation of T cell differentiation | 4.09E-02 |
| GO:0022408 | negative regulation of cell-cell adhesion | 4.71E-02 |
| GO:0050853 | B cell receptor signaling pathway | 4.71E-02 |
Table 5.
Pathways modified in TNFi non-responders at the epigenetic and transcriptomic levels.
| ID | Description | P.adj |
|---|---|---|
| GO:0031098 | stress-activated protein kinase signaling cascade | 2.92E-04 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 3.14E-04 |
| GO:0051403 | stress-activated MAPK cascade | 3.14E-04 |
| GO:0007254 | JNK cascade | 8.28E-03 |
| GO:0002718 | regulation of cytokine production involved in immune response | 1.20E-02 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1.20E-02 |
| GO:0046328 | regulation of JNK cascade | 1.62E-02 |
| GO:0002699 | positive regulation of immune effector process | 1.76E-02 |
| GO:0002285 | lymphocyte activation involved in immune response | 3.65E-02 |
| GO:0002456 | T cell mediated immunity | 3.78E-02 |
| GO:0030217 | T cell differentiation | 4.69E-02 |
List of 73 pathways significantly modified by TNFi therapy in responder patients between week0 and week 12 at both the methylation and gene expression levels. ID: Gene Ontology id; Description: biological annotation of the gene ontology; P.adj: FDR-adjusted significance level for pathway association at the transcriptomic level.
List of 11 pathways significantly modified by TNFi therapy in non-responder patients between week0 and week 12 at both the methylation and gene expression levels. ID: Gene Ontology id; Description: biological annotation of the gene ontology; P.adj: FDR-adjusted significance level for pathway association at the transcriptomic level
Discussion
Defining the factors mediating the response to TNFi therapy is one of the main challenges of precision medicine in RA. Using an epigenome-wide longitudinal design, we have been able to demonstrate that treatment with TNFi changes the blood methylation state of RA patients towards that of healthy individuals. We have also found that the methylation variation in pathways associated with T cell activation, GTPase signaling and actin cytoskeleton changes detected at baseline is associated with the clinical response three months later. Using cell-deconvolution analysis, we have found that drug response is associated with methylation variation in different immune cells, with monocytes displaying a predominant role. The results of this study show that epigenetic variation from circulating immune cells is useful to understand the heterogeneity of response to TNFi.
There is currently little understanding of the association between the changes occurring at the clinical level and those occurring at the epigenetic level. To our knowledge, our study is the first to analyze the methylation changes induced by this commonly used targeted therapy in RA. The longitudinal analysis of methylation in RA patients in this study demonstrates that the systemic blocking of TNF has a significant impact on the biological functions that are associated with disease. We also show that these methylation changes systematically move towards the methylation state of healthy individuals. In line with recent work evaluating the response to TNFi at the transcriptomic level35, we have found that TNFi non-responders show numerous changes in their circulating epigenome (n=57 pathways, FDR<0.05) albeit less abundant compared to treatment responders (n=99 pathways, FDR<0.05). Most of the pathways modified by TNFi in responders were also modified in non-responders (n=45, 78.9%), suggesting that TNF inhibition has a large impact in the immune system irrespective of the clinical outcome. Importantly, we have also found a significant overlap between the functional changes occurring at the epigenetic level with those at the transcriptional level, confirming the presence of a substantial regulatory coupling induced by TNFi therapy in the blood of RA patients.
Although TNF blocking might modify the circulating epigenome in all RA patients, we show that this occurs differently in responders and non-responders. In the longitudinal analysis, we found that 56 pathways that are linked to RA show a different temporal change between responders and non-responders after 12 weeks of therapy. Half of these pathways (51.8%) are also altered by TNFi, suggesting that the magnitude of the epigenetic modification is also an important contributor to the observed clinical differences between responders and non-responders. Among the biological processes that were exclusively modified in responder patients, B cell activation showed the highest significance (FDR = 7e−7). Within non-responders, the regulation of cell killing was found to be the most significantly modified biological activity, which was also exclusive to this group (FDR=5.5e−3). Gene expression data from previous studies corroborated the exclusive longitudinal association of B cell activation in responders (FDR=0.017, Supplementary Table 17), and showed a trend for cell killing activation in non-responders, although it did not reach statistical significance (FDR=0.072, Supplementary Table 18). These results support the presence of an underlying immunological heterogeneity in RA patients that respond differently to the therapeutic suppression of TNF signaling that is detectable in blood.
Of all the biological processes modified by TNFi in both responders and non-responders, signal transduction via small GTPase was found to be the most significantly associated pathway. Small GTPases are a large family of monomeric GTP-binding proteins that are involved in the relay of different signals.36 They are critical regulators of fundamental aspects of activities of cells of the innate and adaptive immune systems, including adhesion and migration.37 The Ras and Rho family of intracellular proteins are among the most well characterized. The latter have been associated to the regulation of the actin cytoskeleton in response to signaling. During APC priming of T cells, Rho-family GTPases like Cdc42 are activated and induce a downstream process that leads to actin polymerization. Without this cytoskeleton reorganization, interactions between the T cell and the APC would not stabilize and T cell activation would completely fail. Similarly, B cell activation requires the participation of small GTPases.38 Small GTPases are essential also for the motility and chemotaxis of monocytes39 and neutrophils.40 Based on this evidence, our results support that the therapeutic dampening of systemic TNF levels has a profound modification of the effector state of most circulating immune cells.
Analysis of the epigenetic profile of RA-linked pathways before treatment initiation showed that TNFi responders have already a different methylation status than non-responders. This significant and reproducible differential methylation involves T cell activation and differentiation, small GTPase signaling and actin filament organization pathways. This result suggests that the epigenetic program of circulating cells conditions the future efficacy of TNF inhibition. Apart from the expected large overlap of genes between T cell activation and differentiation processes, there is very low overlap with the other two biological processes (e.g. 14% gene overlap between actin filament organization and small GTPase signaling). This result suggests that, while these are biological processes that can be strongly linked (e.g. during T cell priming by APCs), they influence response to therapy independently. Among these enriched pathways, differential methylation in the GTPase signaling pathway between responders and non-responders was found to persist after 12 weeks of therapy. Furthermore, this pathway was also associated with drug response at the transcriptional level in a previous study conducted by Tasaki et al. (P<0.05).35 Altogether, these results support the importance of GTPase-dependent immune signal relay in mediating the response to TNFi, and may potentially be a valuable tool to overcome patient heterogeneity and predict clinical outcome.
Using a novel deconvolution method, CellDMC, we have found significant and reproducible methylation variation in specific immune cell types associated with the response to TNFi therapy. CellDMC has been shown to reliably detect cell-specific differential methylation, including B-cell specific DMPs in RA, epithelial-specific DMPs in breast cancer20 or, more recently, cell-specific methylation associated with smoking.29 Like other tissues in the body, blood is a mixture of different cell types. Epigenetics is a key mechanism for cell differentiation and, therefore, each cell type will contribute to specific methylation features.20 This entails that the analysis of whole blood DNA might mask some of the cell-specific epigenetic variation. Deconvolution approaches allow us to identify part of these cell-specific changes. Compared to whole blood, site-specific methylation changes were significant even after multiple testing correction. This result supports the notion that analyzing epigenome-wide methylation in a cell-specific manner can increase statistical power as means to observe biologically relevant alterations.18 Using this approach, cells from the innate and adaptive immune systems were found to be associated with the response to TNFi therapy. The association of differentially methylated sites in CD4+ T cells is in accordance with the findings at the pathway level. Cell deconvolution analysis also showed an important role of innate immune cells in determining therapy response, including NK, neutrophils and monocytes. This result supports that the response to TNFi is the result of the concerted action of different immune cells and does not reside exclusively in a single cell type.
Among the cell types associated with the response to TNFi therapy at baseline, monocytes showed the largest epigenetic differences. This is in accordance with previous studies evaluating methylation on circulating cells in RA. In the largest-scale study of epigenetic variation in RA against controls,16 monocytes were found to contribute to most of the methylation changes occurring in blood. More recently, epigenome analysis of isolated monocytes in RA has identified promising association between CpG variation and disease activity in RA.17 Analysis of these methylation regions in our patient cohort, however, did not show an association with TNFi response (Supplementary Table 19). This result suggests that methylation variation associated with disease activity might be a downstream effect of the global proinflammatory cues in RA, whereas the methylation variation associated to therapy that we observed in our study is more directly associated to the underlying disease heterogeneity. Motif enrichment analyses indicated that hyper- and hypomethylation patterns in monocytes are associated with immune-relevant transcription factor activities, in which these results could have potential therapeutic implications. STAT1 was found to be associated with hypermethylation patterns in responders to TNFi, suggesting that non-responders have a higher level of activation of this pathway. Consequently, therapies aimed at JAK/STAT signaling inhibition that involve STAT1, like oral JAK inhibitor tofacitinib,41 could have a higher probability of success in this latter group of patients. Forthcoming studies evaluating the molecular changes in blood of JAK inhibitor therapies in RA will be of high value to corroborate the utility of our findings for patient therapeutic stratification.
The identification of epigenetic variation in blood associated with TNFi response is of high relevance in RA. Two previous studies have been performed to date with this objective. In the first study,31 whole blood methylation was analyzed at baseline from a cohort of patients starting treatment with etanercept, a fusion protein of p75 TNF receptor and the Fc fragment of IgG1. Five methylation sites were associated with the response to therapy. In our data, none of the CpGs showed a differential methylation with TNFi response at the nominal level, even after stratifying for drug type. In a more recent study,32 an opposing methylation profile of nominally associated CpGs (P<0.05) was found in the peripheral blood mononuclear cells (PBMCs) at baseline from patients treated with adalimumab (a fully humanized monoclonal anti-TNF antibody) and etanercept. This pattern was not validated in the two RA cohorts analyzed in our study. There are different possible explanations for the lack of reproducibility for these previous findings, including the exclusion of the methylation profile of the granulocyte fraction (the most abundant cell type in blood) in PBMC data and the low number of non-responders compared to responders. However, we argue that this lack of consistency might be largely due to a general lack of statistical power of epigenetic studies in blood. In contrast to our study, none of the previous studies performed an independent validation. This was possible due to the use of more powerful analysis strategies, such as pathway analysis and cell type deconvolution. Hence, together with validation in an independent patient cohort, we were able to discover and validate new epigenetic features associated with the response to TNFi therapy.
Nevertheless, our study presents several limitations. First, lack of statistical power of small cohorts may impede the discovery of other biologically relevant pathways that contribute to TNFi response or the study of clinical subgroups of patients. Second, further studies are required to fully understand the mechanisms behind the methylation variation that we identified at baseline in order to predict patient response to TNFi. This is particularly relevant in the case of cell-specific DMPs. The pathway-based analysis provides a more readily interpretation, but the cell specific methylation does not directly translate in many cases into a specific biological function. While genes like ALOX5 or MAML3 have been found to be highly expressed in the associated cell types (neutrophils and monocytes, respectively), the relation of other associated genes with the specific cell type is less clear and will require additional work. Third, the evaluation of methylation data later time points (e.g. weeks 16-26, week 48) might have likely increased the power to identify the most consistent epigenetic changes associated with the clinical response. Finally, our cell type deconvolution analysis only allowed us to identify the contribution of a the main immune cell lineages, while it has been described that specific cell subtypes, such as monocyte-derived cells including osteoclasts42 and dendritic cells,43 as well as various T cell subsets including Th1, Th2 and Th17,44 are implicated in joint destruction and response to treatment.
In conclusion, using a longitudinal study of the blood methylome, we have identified the dynamic biological changes associated with TNFi therapy. We have also found that pathways that are associated with RA are also associated with the clinical response to therapy at baseline. Using cell-deconvolution analysis we show that the response to TNF involves different immune cell types, with monocytes being at the apex of this patient diversity. The present work sets the basis for the development of epigenetic-based biomarkers for patient stratification and provide support for alternative, more efficacious therapies for TNFi non-responders.
Contributors
AJ and SM designed the study, and wrote the initial draft of the manuscript. AG, SHMM, TL and AJ performed data analysis. DA and RM performed the DNA methylation microarray assays. Patient recruitment and in depth-clinical characterization of the two longitudinal cohorts was performed by MLL, FB, AE, AFN, AJM, CPG, MLGV, SSF, MAL, RS, AMO, CMFC, CDT, EM, JTM and SM. All authors participated manuscript draft or critical revision, and contributed significantly to acquisition and analysis of the data and interpretation of the results.
Data sharing
DNA methylation data is available for download from Gene Expression Omnibus database with accession number GSE176168 .
Declaration of interests
SM and RMM are co-founders of IMIDomics, Inc. AJ is Chief Data Scientist of IMIDomics, Inc. The remaining authors declare that they have no conflict of interest.
Acknowledgements
We thank the RA patients participating in the study and the clinical personnel from the IMID Consortium for their participation. This study was funded by the Instituto de Salud Carlos III.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104053.
Contributor Information
Antonio Julià, Email: toni.julia@vhir.org.
Sara Marsal, Email: sara.marsal@vhir.org.
Appendix. Supplementary materials
References
- 1.Myasoedova E., Crowson C.S., Kremers H.M., Therneau T.M., Gabriel S.E. Is the incidence of rheumatoid arthritis rising?: results from Olmsted County, Minnesota, 1955–2007. Arthritis Rheum. 2010;62(6):1576–1582. doi: 10.1002/art.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felson D.T., Smolen J.S., Wells G., et al. American College of Rheumatology/European League against Rheumatism provisional definition of remission in rheumatoid arthritis for clinical trials. Ann Rheum Dis. 2011;70(3):404–413. doi: 10.1136/ard.2011.149765. Mar. [DOI] [PubMed] [Google Scholar]
- 3.Pitzalis C., Choy E.H., Buch M.H. Transforming clinical trials in rheumatology: towards patient-centric precision medicine. Nat Rev Rheumatol. 2020;16(10):590–599. doi: 10.1038/s41584-020-0491-4. [DOI] [PubMed] [Google Scholar]
- 4.Bergman M.J., Kivitz A.J., Pappas D.A., et al. Clinical utility and cost savings in predicting inadequate response to Anti-TNF therapies in rheumatoid arthritis. Rheumatol Ther. 2020;7(4):775–792. doi: 10.1007/s40744-020-00226-3. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor P.C., Feldmann M. Anti-TNF biologic agents: still the therapy of choice for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5(10):578–582. doi: 10.1038/nrrheum.2009.181. [DOI] [PubMed] [Google Scholar]
- 6.Lipsky P.E., van der Heijde D.M., St. Clair E.W., et al. Infliximab and methotrexate in the treatment of rheumatoid arthritis. N Engl J Med. 2000;343(22):1594–1602. doi: 10.1056/NEJM200011303432202. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Mola E., Balsa A., García-Vicuna R., et al. Anti-citrullinated peptide antibodies and their value for predicting responses to biologic agents: a review. Rheumatol Int. 2016;36(8):1043–1063. doi: 10.1007/s00296-016-3506-3. [DOI] [PubMed] [Google Scholar]
- 8.Acosta-Colman I., Palau N., Tornero J., et al. GWAS replication study confirms the association of PDE3A–SLCO1C1 with anti-TNF therapy response in rheumatoid arthritis. Pharmacogenomics. 2013;14(7):727–734. doi: 10.2217/pgs.13.60. [DOI] [PubMed] [Google Scholar]
- 9.Julià A., Erra A., Palacio C., et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4(10):e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lequerré T., Gauthier-Jauneau A.C., Bansard C., et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8(4):1–11. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiguchi N., Kawauchi S., Furuya T., et al. Messenger ribonucleic acid expression profile in peripheral blood cells from RA patients following treatment with an anti-TNF-α monoclonal antibody, infliximab. Rheumatology. 2008;47(6):780–788. doi: 10.1093/rheumatology/ken083. [DOI] [PubMed] [Google Scholar]
- 12.Kim M., Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp Mol Med. 2017;49(4):e322. doi: 10.1038/emm.2017.10. Apr–e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karouzakis E., Raza K., Kolling C., et al. Analysis of early changes in DNA methylation in synovial fibroblasts of RA patients before diagnosis. Sci Rep. 2018;8(1):7370. doi: 10.1038/s41598-018-24240-2. May 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ai R., Whitaker J.W., Boyle D.L., et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 2015;67(7):1978–1980. doi: 10.1002/art.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Yim A.Y.F., Ferrero E., Maratou K., et al. Novel insights into rheumatoid arthritis through characterization of concordant changes in DNA methylation and gene expression in synovial biopsies of patients with differing numbers of swollen joints. Front Immunol. 2021;12:1215. doi: 10.3389/fimmu.2021.651475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Aryee M.J., Padyukov L., et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez-Ubreva J., la C.F.C., Li T., et al. Inflammatory cytokines shape a changing DNA methylome in monocytes mirroring disease activity in rheumatoid arthritis. Ann Rheum Dis. 2019;78(11):1505–1516. doi: 10.1136/annrheumdis-2019-215355. Nov 1. [DOI] [PubMed] [Google Scholar]
- 18.Ballestar E., Li T. New insights into the epigenetics of inflammatory rheumatic diseases. Nat Rev Rheumatol. 2017;13(10):593–605. doi: 10.1038/nrrheum.2017.147. [DOI] [PubMed] [Google Scholar]
- 19.Glossop J.R., Nixon N.B., Emes R.D., et al. DNA methylation at diagnosis is associated with response to disease-modifying drugs in early rheumatoid arthritis. Epigenomics. 2017;9(4):419–428. doi: 10.2217/epi-2016-0042. Apr 1. [DOI] [PubMed] [Google Scholar]
- 20.Zheng S.C., Breeze C.E., Beck S., Teschendorff A.E. Identification of differentially methylated cell types in epigenome-wide association studies. Nat Methods. 2018;15(12):1059–1066. doi: 10.1038/s41592-018-0213-x. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aterido A., Cañete J.D., Tornero J., et al. Genetic variation at the glycosaminoglycan metabolism pathway contributes to the risk of psoriatic arthritis but not psoriasis. Ann Rheum Dis. 2019;78(3):355–364. doi: 10.1136/annrheumdis-2018-214158. [DOI] [PubMed] [Google Scholar]
- 22.Fransen J., Van Riel P. The disease activity score and the EULAR response criteria. Rheum Dis Clin North Am. 2009;35(4):745–757. doi: 10.1016/j.rdc.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Rubbert-Roth A., Enejosa J., Pangan A.L., et al. Trial of upadacitinib or abatacept in rheumatoid arthritis. N Engl J Med. 2020;383(16):1511–1521. doi: 10.1056/NEJMoa2008250. [DOI] [PubMed] [Google Scholar]
- 24.Triche Jr T.J., Weisenberger D.J., Van Den Berg D., Laird P.W., Siegmund K.D. Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090. –e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie M.E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. Apr 20–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phipson B., Maksimovic J., Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–288. doi: 10.1093/bioinformatics/btv560. Jan 15. [DOI] [PubMed] [Google Scholar]
- 27.Maksimovic J., Gagnon-Bartsch J.A., Speed T.P., Oshlack A. Removing unwanted variation in a differential methylation analysis of Illumina HumanMethylation450 array data. Nucleic Acids Res. 2015;43(16):e106. doi: 10.1093/nar/gkv526. Sep 18–e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maksimovic J., Oshlack A., Phipson B. Gene set enrichment analysis for genome-wide DNA methylation data. Genome Biol. 2021;22(1):1–26. doi: 10.1186/s13059-021-02388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You C., Wu S., Zheng S.C., et al. A cell-type deconvolution meta-analysis of whole blood EWAS reveals lineage-specific smoking-associated DNA methylation changes. Nat Commun. 2020;11(1):4779. doi: 10.1038/s41467-020-18618-y. Sep 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houseman E.A., Accomando W.P., Koestler D.C., et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plant D., Webster A., Nair N., et al. Differential methylation as a biomarker of response to etanercept in patients with rheumatoid arthritis. Arthritis Rheumatol. 2016;68(6):1353–1360. doi: 10.1002/art.39590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao W., Concepcion A.N., Vianen M., et al. Multiomics and machine learning accurately predict clinical response to adalimumab and etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73(2):212–222. doi: 10.1002/art.41516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plant D., Prajapati R., Hyrich K.L., et al. Replication of association of the PTPRC gene with response to anti–tumor necrosis factor therapy in a large UK cohort. Arthritis Rheum. 2012;64(3):665–670. doi: 10.1002/art.33381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julià A., Ávila G., Celis R., et al. Lower peripheral helper T cell levels in the synovium are associated with a better response to anti-TNF therapy in rheumatoid arthritis. Arthritis Res Ther. 2020;22(1):196. doi: 10.1186/s13075-020-02287-9. Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tasaki S., Suzuki K., Kassai Y., et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Masri R., Delon J. RHO GTPases: from new partners to complex immune syndromes. Nat Rev Immunol. 2021:1–15. doi: 10.1038/s41577-021-00500-7. [DOI] [PubMed] [Google Scholar]
- 37.van H.S.F.G., EC A., Dee R., Hordijk P.L. Rho GTPase expression in human myeloid cells. PLOS ONE. 2012;7(8):e42563. doi: 10.1371/journal.pone.0042563. Aug 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saci A., Carpenter C.L. RhoA GTPase regulates B cell receptor signaling. Mol Cell. 2005;17(2):205–214. doi: 10.1016/j.molcel.2004.12.012. Jan 21. [DOI] [PubMed] [Google Scholar]
- 39.Jones G.E., Allen W.E., Ridley A.J. The Rho GTPases in macrophage motility and chemotaxis. Cell Adhes Commun. 1998;6(2–3):237–245. doi: 10.3109/15419069809004479. [DOI] [PubMed] [Google Scholar]
- 40.Tackenberg H., Möller S., Filippi M.D., Laskay T. The Small GTPase Cdc42 is a major regulator of neutrophil effector functions. Front Immunol. 2020;11:1197. doi: 10.3389/fimmu.2020.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle D., Soma K., Hodge J., et al. The JAK inhibitor tofacitinib suppresses synovial JAK1-STAT signalling in rheumatoid arthritis. Ann Rheum Dis. 2015;74(6):1311–1316. doi: 10.1136/annrheumdis-2014-206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue J., Xu L., Zhu H., et al. CD14+ CD16- monocytes are the main precursors of osteoclasts in rheumatoid arthritis via expressing Tyro3TK. Arthritis Res Ther. 2020;22(1):1–11. doi: 10.1186/s13075-020-02308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marzaioli V., Canavan M., Floudas A., et al. CD209/CD14+ dendritic cells characterization in rheumatoid and psoriatic arthritis patients: activation, synovial infiltration, and therapeutic targeting. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.722349. –722349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulic S., Vásárhelyi Z., Sava F., et al. T-cell subsets in rheumatoid arthritis patients on long-term anti-TNF or IL-6 receptor blocker therapy. Mediators Inflamm. 2017:2017. doi: 10.1155/2017/6894374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.