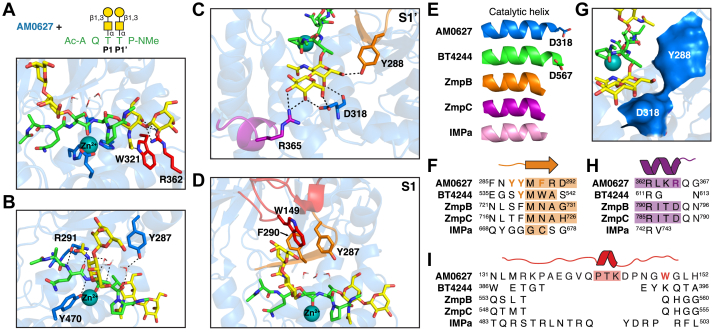
Figure 3.
Molecular modeling reveals residues involved in the recognition of adjacent truncated O-glycans.A, docking of glycopeptide Ac-AQT∗T∗P-NMe into AM0627, where asterisks indicate glycosylated residues. Gluzincin motif residues are shown as blue sticks and S1′ subsite G1′ GalNAc-binding residues are shown as red sticks. B, glycopeptide backbone contacts with AM0627 and inserted active site water molecules. C and D, nonconserved and semiconserved S1’ (C) and S1 (D) subsite glycan-interacting residues. In (A–D), the Zn2+ ion is represented as a teal sphere and hydrogen bonds are depicted as black dashes. E, comparison of catalytic helices between PF13402 enzymes. The spatially conserved aspartate residues in AM0627 and BT4244 are shown as sticks. F, active site beta strand sequence alignments between PF13402 enzymes determined using the DALI server (31). G, surface representation of residues Y288 and D318 in the S1′ subsite. H, G2’’ glycan-binding S1′ subsite sequence alignments between PF13402 enzymes determined using the DALI server. I, S1 subsite loop sequence alignments between PF13402 enzymes determined using the DALI server. In (F, H and I), colored regions highlight strand/helix sequences.