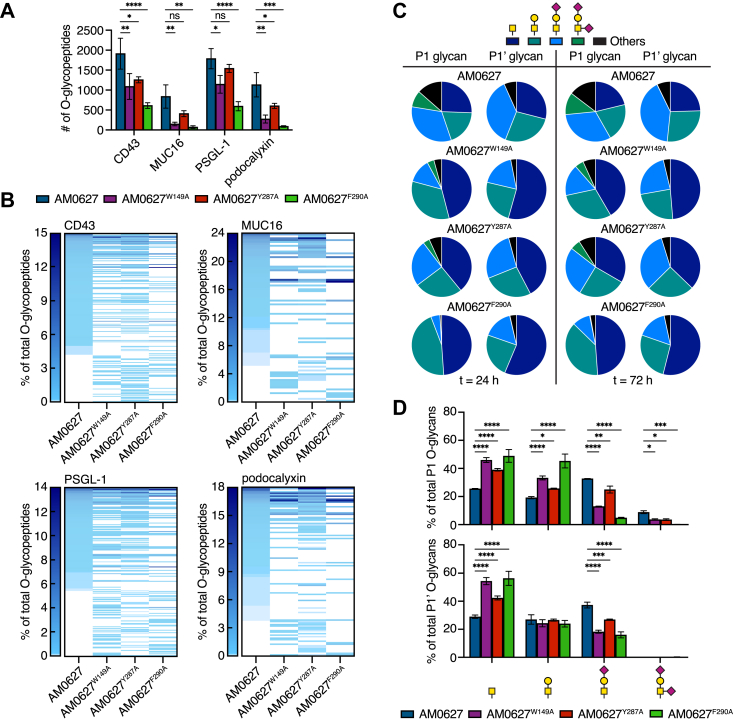
Figure 5.
Mass spectrometry analysis reveals altered substrate preferences.A, total number of generated glycopeptides with high confidence O-glycosite localization for each substrate after a 24-h incubation with AM0627 or point mutants. B, heat maps depicting the frequency of each O-glycopeptide sequence normalized to the total number of substrate O-glycopeptides generated by the enzyme after 24 h. Peptide sequences are ordered from highest to lowest frequency (top to bottom) for AM0627. Specific sequences and counts are listed in Dataset S3. C, distributions of O-glycan occurrences at P1 and P1′ in O-glycopeptides generated by each enzyme after 24- and 72-h digests. D, quantification of O-glycan occurrences at P1 (top) and P1’ (bottom) for 24-h digests. Data are mean ± s.d. (n = 2). p-values were determined by two-way ANOVA with Dunnett correction. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.