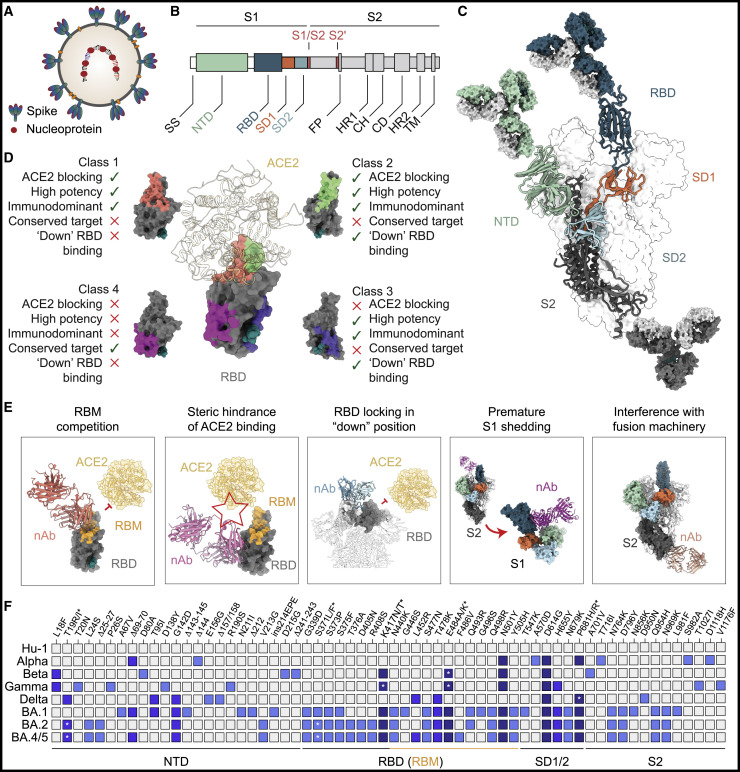
Figure 2.
Targets and mechanisms of SARS-CoV-2 neutralization
(A) Schematic of a SARS-CoV-2 virion indicating the spike and nucleocapsid proteins, the two major targets of SARS-CoV-2-reactive antibodies.
(B) Schematic of the S1/S2 heterodimer domains encoded by the SARS-CoV-2 spike gene. Host protease cleavage sites are indicated in red.
(C) SARS-CoV-2 trimer structure with heterodimer domains colored as in (B) and main targets of neutralizing activity (RBD, NTD, and S2) indicated by bound antibodies.
(D) Exemplary classification system of RBD neutralizing antibodies as defined in (Barnes et al., 2020a). Key features of different RBD antibody classes are indicated with green ticks (present) or red crosses (absent).
(E) Examples for mechanisms of SARS-CoV-2 neutralization.
(F) Map of spike gene changes in SARS-CoV-2 variants of concern relative to the ancestral Hu-1 strain (changes found in >33% of variant sequences deposited at GISAID (Elbe and Buckland-Merrett, 2017; Khare et al., 2021; Shu and McCauley, 2017) and as aggregated at outbreak.info (Mullen et al., 2020). Changes are colored based on their frequency amongst variants of concern (light blue, 1; blue, 2; dark blue, 3 or more variants with identical change; with Omicron sublineages [BA.1, BA.2, BA.4/BA.5] counted as single variant). Asterisks refer to amino acid polymorphism indicated by corresponding asterisks above. SS, signaling sequence; NTD, N-terminal domain; RBD, receptor-binding domain; SD1/SD2, subdomain 1/2; FP, fusion peptide; HR, heptad repeat; CH, central helix; CD, connector domain; TM, transmembrane domain; RBM, receptor-binding motif; nAb, neutralizing antibody.