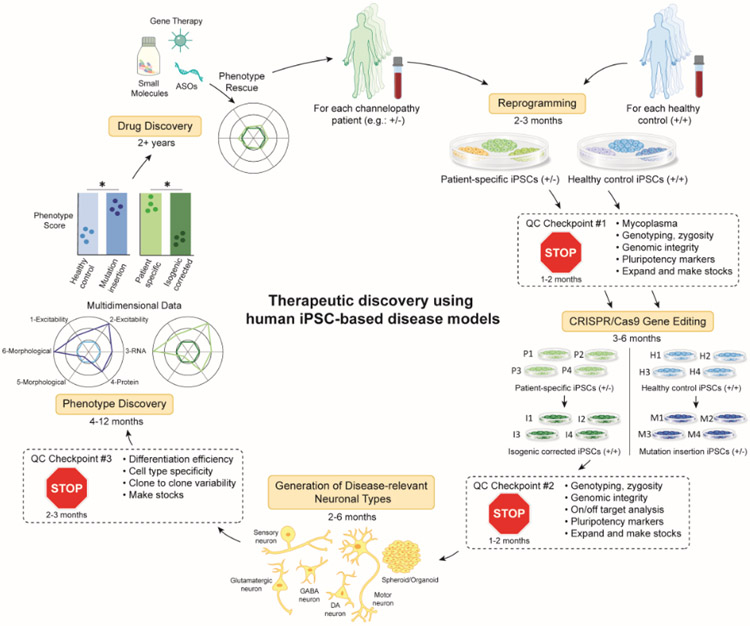
Figure 2. Flowchart of research practices for iPSC disease modeling and therapeutic discovery.
Here we present an example flowchart using a patient, heterozygous for a disease-associated mutation (+/−), although this process can be applied to patients with other types of mutations. Somatic cells obtained from a patient or alternatively a healthy control subject are reprogrammed into iPSCs (See Table S2). Several clones can be screened at QC (Quality Control) checkpoint 1 (See Text box 1). Genetic editing technology, such as CRISPR/Cas9 can be used to generate isogenic controls where a pathogenic variant is corrected in patient-derived lines or inserted in healthy control iPSCs. Several clones of each should be screened at QC checkpoint 2. iPSCs can then be differentiated into disease relevant cell types using 2D or 3D protocols (See Table S3). Each type of differentiation protocol must be optimized and screened at QC checkpoint 3. At this point, in vitro phenotype discovery is initiated in which any number of assays relevant to the disease (functional, morphological, biochemical) are performed and multi-dimensional metrics are compiled into an ultimate phenotype score. Ideally, there should be overlap in the phenotypes scores between patient lines that have been corrected (P->I) and healthy control lines that have been mutated (H->M). Once a robust, reproducible in vitro phenotype is established, drug discovery can be initiated. This workflow should be repeated ideally with multiple patients and healthy controls. The approximate timeline for completing each step is noted throughout the figure.