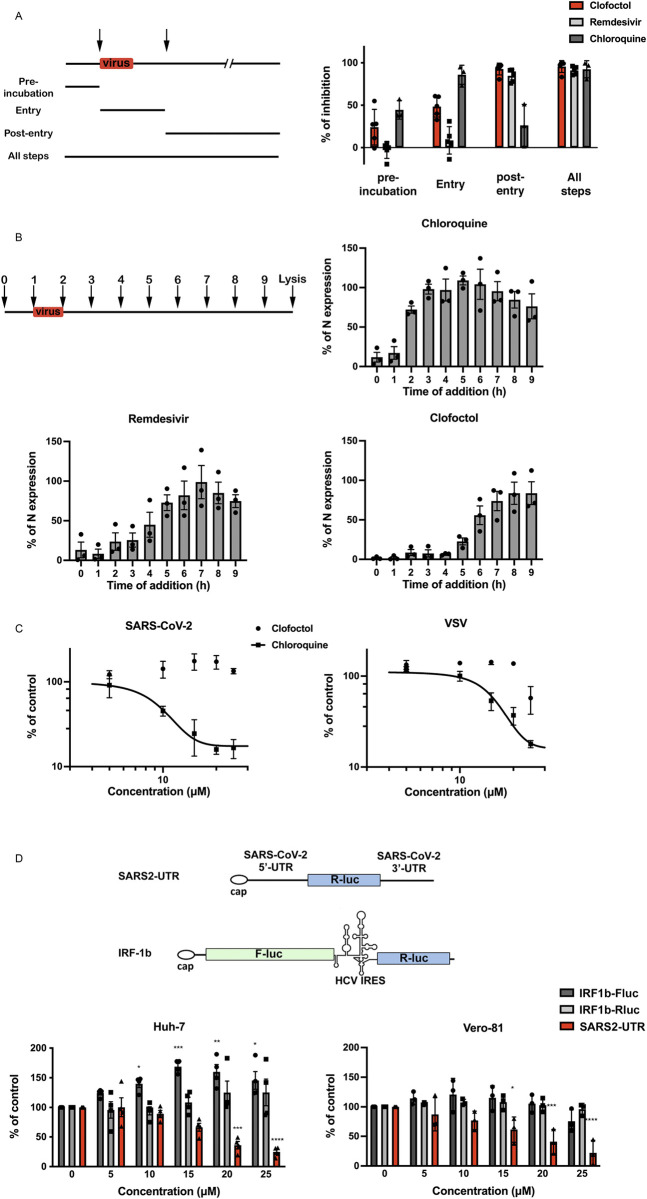
Fig 3. Clofoctol targets the translation step of SARS-CoV-2 life cycle.
A, Clofoctol is mainly efficient at a post-entry step. Vero-81 cells were infected with SARS-CoV-2 at an MOI of 0.25. Clofoctol, remdesivir or CQ were present at a concentration of 15 μM either before infection, during virus entry, post-inoculation or throughout the steps as indicated on the schematic depiction of the experiment. Bars indicate when the drugs are present during the experiment for each condition. At 16h post-infection, cells were fixed with 4% paraformaldehyde and processed to detect the proportion of infected cells. Therefore, they were immunostained to allow for the detection of the viral double-stranded RNA and nuclei were detected by Hoechst staining to count the total number of cells. Results are presented as the percentage of infection inhibition and represent the average of five independent experiments. B, Time-of-addition experiment. Vero-81 cells were infected at an MOI of 0.5 for 1h. 15 μM of clofoctol, remdesivir or CQ were added every hour starting 1h before inoculation. Cells were lysed 8h after the end of the inoculation in Laemmli loading buffer and the amount of N protein was detected in immunoblot. Results are presented as the percentage of N protein expression relative to that in non-treated cells (CTL) and represent the average of three independent experiments. Error bars represent the standard error of the mean (SEM). C, Clofoctol does not inhibit SARS-CoV-2 entry. Huh-7 cells expressing ACE2 receptor were infected with SARS2pp or pseudoparticles containing the envelope glycoprotein of the vesicular stomatitis virus (VSV) used as a control (VSVpp) for 3 hours in the presence of increasing concentrations of clofoctol or CQ. At 48 hours post-infection, cells were lysed to quantify luciferase activity. The results are expressed in % of the controls of three independent experiments. The experiments were performed in triplicate (n = 3) in each condition. D, Clofoctol inhibits viral RNA translation. Schematic representation of the reporter construct expressing the Renilla luciferase placed between the 5’-UTR and the 3’-UTR of the SARS-CoV-2 genomic RNA and the control bicistronic construct containing the firefly luciferase sequence under the control of a cap structure, followed by the Renilla luciferase under the control of hepatitis C virus (HCV) IRES. Huh-7 or Vero-81 cells were electroporated with in vitro transcribed RNA. Cells were lysed after 8h and luciferase activities were recorded. The results are expressed in % of the controls of three independent experiments. The experiments were performed in quadruplicate (n = 4) in each condition. Two-way ANOVA followed by the Dunnett’s multiple comparisons test was performed for statistical analysis (*p < 0.05; **p < 0.01; ***p < 0.001).