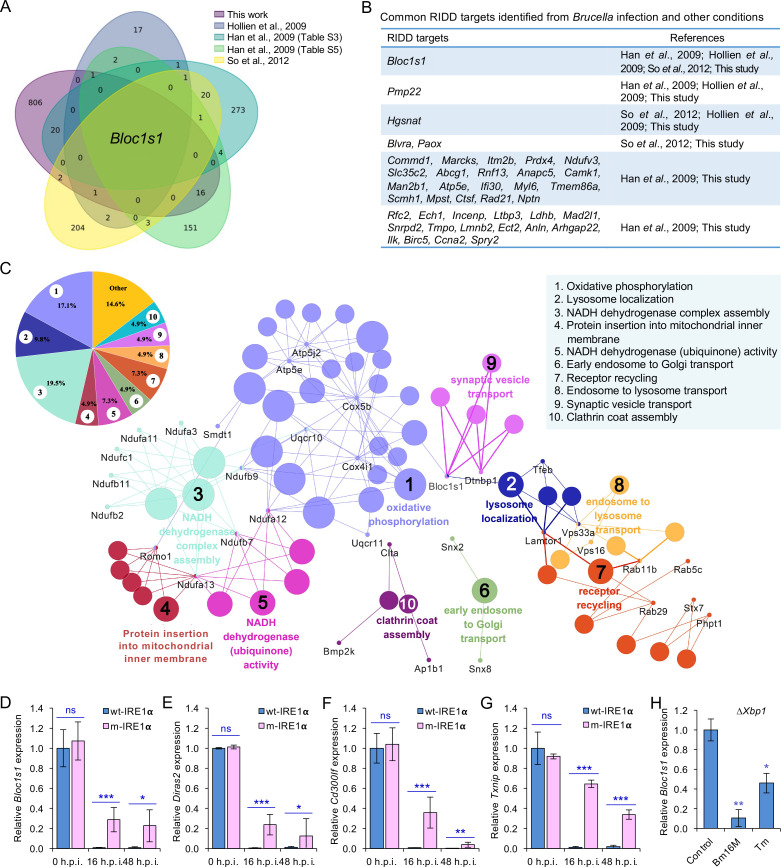
Figure 3. Identification of host RIDD targets during Brucella infection.
(A) Venn diagram showing numbers of candidate RIDD genes identified in the indicated datasets. (B) Common candidate RIDD genes identified in Bm16M-infected cells and other conditions in the indicated datasets. (C) Interaction network analysis of candidate RIDD genes (Bloc1s1 associated genes) identified in Brucella-infected cells and the corresponding enriched KEGG pathways. Different pathways are distinguished by different colors. Interacting genes are shown with the smallest sized of dot with gene names. The upper-left-corner panel: enriched KEGG pathways and interacting candidate RIDD genes (%). qRT-PCR validation of RIDD candidate genes Bloc1s1 (D), Diras2 (E), Cd300lf (F), and Txnip (G) identified from RNA-seq analysis. Relative mRNA expression levels in potential RIDD targets from control and m-IRE1α BMDMs infected with Bm16M at 16 and 48 h.p.i. were measured by qRT-PCR. (H) qRT-PCR analysis of expression levels of Bloc1s1 in ΔXbp1 BMDMs that were either uninfected (control), infected with Bm16M, or treated with tunicamycin (Tm, an UPR inducer, 5 μg/ml) at 4-hr post infection/treatment. Expression levels of the indicated genes were normalized to Gapdh expression. Statistical data represent the mean ± standard error of mean (SEM) from three independent experiments. *, **, and ***: significance at p < 0.05, 0.01, and 0.001, respectively.