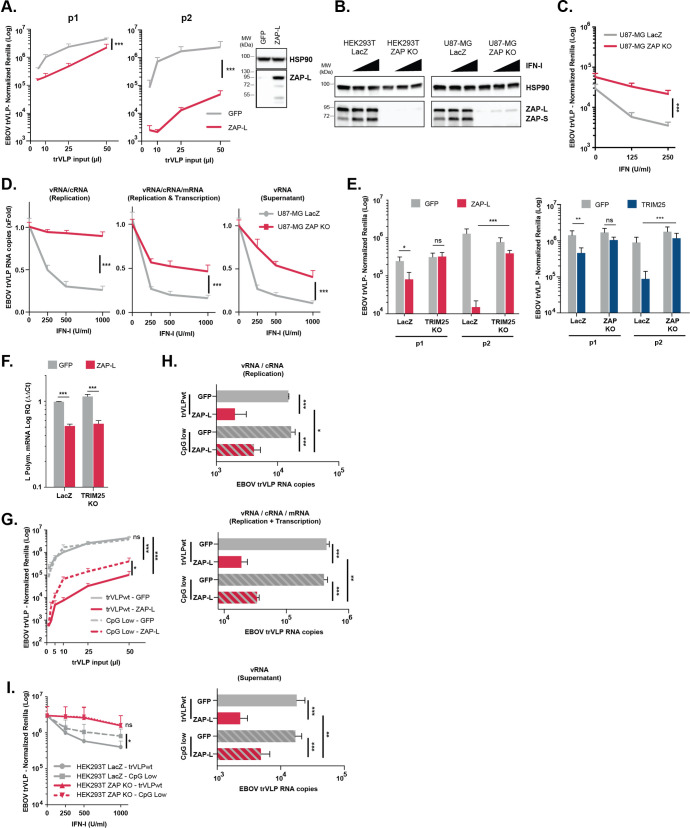
Fig 4. TRIM25 and ZAP are inter-dependent for their antiviral activity against EBOV trVLP.
(A) EBOV trVLP normalized reporter activity on HEK293T-TIM1 cells transfected with EBOV RNP proteins and either GFP (grey) or ZAP-L (red) prior to infection with increasing amounts of EBOV trVLPs (p1 target cells, left panel). Supernatants from p1 cells were harvested and used to infect p2 target cells (middle panel), and reporter activities measured 24 hours later. Protein levels of HSP90 and ZAP were determined by western blot at time of infection. (B) HEK293T- and U87-MG-based CRISPR cells lines were treated with increasing amounts of IFN-I, and lysed 24 hours later for analysis. Protein levels of HSP90 and ZAP were determined by western blot on LacZ CRISPR control cells and corresponding ZAP CRISPR KO cell lines. (C) EBOV trVLP reporter activities on U87-MG LacZ CRISPR-TIM1 (grey) and U87-MG ZAP CRISPR KO-TIM1 (red) target cells (p1), transfect with RNP proteins and pre-treated with increasing amounts of IFN-I prior to infection. Reporter activities measured 24 hours after infection. (D) Quantification of intracellular and supernatant viral RNA levels on U87-MG LacZ CRISPR-TIM1 (grey) and U87-MG ZAP CRISPR KO-TIM1 target cells (red) that were transfected with EBOV RNP proteins and pre-treated with IFN-I, prior to infection with EBOV trVLPs as in (C). Random hexamer primers were used to generate cDNAs and qPCR analysis was performed using primers/probe sets targeting either the trailer region of the viral genome (vRNA and cRNA, left panel), or VP40 RNA (intracellular vRNA, cRNA and mRNA, middle panel; minigenomic RNA in the supernatant, right panel). Data presented as fold change compared to control (no IFN) based on absolute copy numbers. (E) EBOV trVLP normalized reporter activities on HEK293T LacZ CRISPR, HEK293T TRIM25 CRISPR KO and HEK293T ZAP CRISPR KO cells stably expressing TIM1, that were transfected with EBOV RNP plasmids together with GFP (grey), ZAP L (red) or TRIM25 (blue), prior to infection with a fixed amount of EBOV trVLPs (p1 target cells). Supernatants from p1 were then harvested and used to infect HEK293T-TIM1 cells (p2 target cells). Protein levels of HSP90, TRIM25 and ZAP at time of infection shown in S4D Fig. (F) Relative quantification of EBOV L-Polymerase RNA transcripts on cell lysates of HEK293T LacZ CRISPR-TIM1 and HEK293T TRIM25 CRISPR KO-TIM1 cells transfected EBOV RNP plasmids in combination with either with GFP (grey) or ZAP-L (red), and infected with a fixed amount of EBOV trVLPs. Random hexamer primers were used to generate cDNAs from total RNA, and RT-qPCR analysis performed using a primers/probe sets targeting EBOV L-polymerase and gapdh. Data normalized to L-polymerase RNA levels on HEK293T LacZ CRISPR-TIM1 cells transfected with GFP based on ΔΔCt values. (G) EBOV trVLP normalized reporter activity on p2 target cells. HEK293T-TIM1 p1 cells were transfected with EBOV RNP proteins, and either GFP (grey) or ZAP-L (red) prior to infection with increasing amounts of wild-type EBOV trVLPs (trVLPwt, solid lines) or a variant with no CpG dinucleotides on the Renilla ORF of the 4cis genome (CpG low, dashed lines). Supernatants from p1 were harvested and used to infect HEK293T-TIM1 p2 target cells. (H) Quantification of viral RNA transcripts present intracellularly (upper and middle panels) and in supernatants (lower panel) of HEK293T-TIM1 cells transfected as in (G), and infected with a fixed amount of EBOV trVLP WT or CpG low. Random hexamer primers were used to generate cDNAs from total RNA and RT-qPCR analysis was performed as in (D). (I) EBOV trVLP normalized reporter activity on HEK293T-TIM1 p2 target cells. HEK293T LacZ CRISPR-TIM1 (grey) and HEK293T ZAP CRISPR KO-TIM1 cells (red) were transfected with EBOV RNP proteins and pre-treated with increasing amounts of IFN-I prior to infection with pre-determined and equivalent amount (1x106 RLU) of wild-type EBOV trVLPs (solid lines) or CpG Low EBOV trVLP (dashed lines). Supernatants from p1 were harvested and used to infect HEK293T-TIM1 p2 target cells. Reporter activities measured 24 hours post-infection. All the represented EBOV trVLP Renilla reporter activities are normalized to control Firefly luciferase values obtained in the same lysates. P > 0.05, **p > 0.01 and ***p > 0.001 as determined by two-tailed paired t-test. All error bars represent ± SEM of at least three independent experiments.