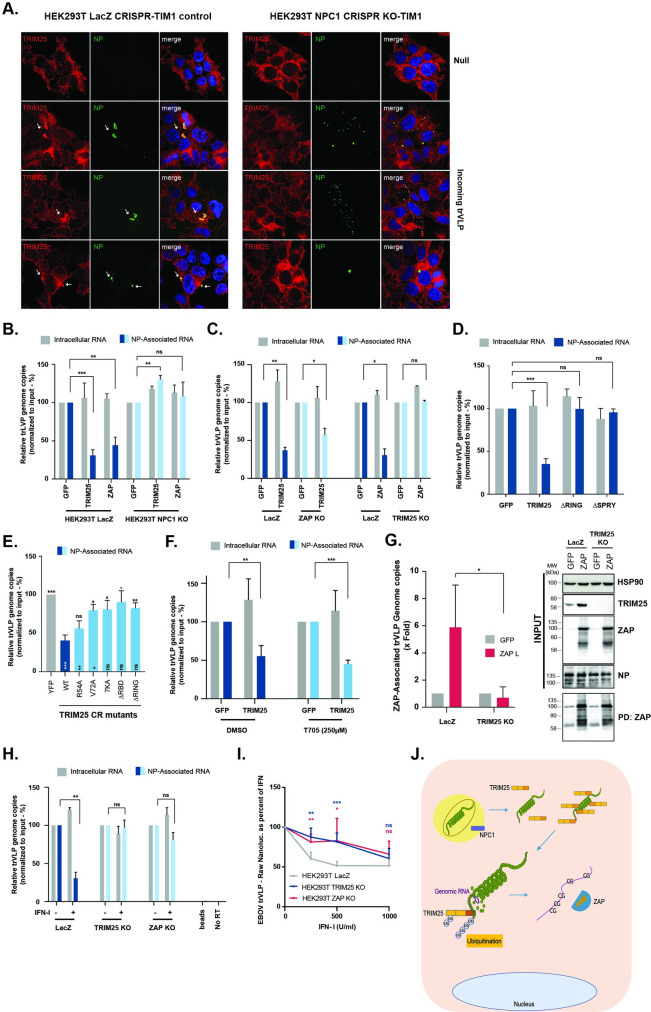
Fig 7. TRIM25 and ZAP promote the dissociation of EBOV trVLP genomic RNA from the viral ribonucleoprotein.
(A) Typical confocal microscopy fields from HEK293T LacZ CRISPR-TIM1 (left panels) or HEK293T NPC1 CRISPR KO-TIM1 cells (right panels) left untreated (Null) or infected with EBOV trVLPs concentrated on a 20% sucrose-cushion. Cells were stained 4 to 6 hours post-infection with anti-TRIM25 (red), anti-EBOV NP (green) and DAPI (blue). White arrows show localization of TRIM25 intracellular aggregates. (B) Relative quantification of intracellular RNA levels (grey) and NP-associated RNA (blue) on HEK293T LacZ CRISPR-TIM1 and HEK293T NPC1 CRISPR KO-TIM1 cells transfected with GFP, TRIM25 or ZAP-L prior to infection with EBOV trVLPs. 3 hours post-infection cells were UV cross-linked, and EBOV NP from incoming virions were immunoprecipitated from lysates with an anti-NP antibody. Following proteinase K treatment, pulled-down RNA was extracted with Qiazol / chloroform, and random hexamer primers were used to generate cDNAs, and qPCR analysis performed using a primers/probe set targeting EBOV VP40 RNA. Values are presented as percentage of absolute RNA copy numbers on cells transfected with GFP. (C) HEK293T LacZ CRISPR, HEK293T ZAP CRISPR KO and HEK293T TRIM25 CRISPR KO cells stably expressing TIM1 were transfected with GFP, TRIM25 or ZAP-L as depicted in the panels, and later infected with EBOV trVLPs. Relative quantification of intracellular viral RNA levels (grey) and NP-associated RNA (coloured bars) were determined as in (B). (D) HEK293T-TIM1 cells were transfected with GFP, wild-type TRIM25 or mutants thereof prior to infection with EBOV trVLPs. Relative quantification of intracellular viral RNA levels (grey) and NP-associated RNA (blue) were determined as in (B). (E) HEK293T TRIM25 CRISPR KO-TIM1 cells were transfected with GFP (grey), or CRISPR-resistant versions of TRIM25 (wild-type, dark blue; or mutants thereof, as depicted in the figure, light blue) prior to infection with EBOV trVLPs. Relative quantification of NP-associated RNA was determined as in (B). (F) Prior to infection with EBOV trVLPs, HEK293T LacZ CRISPR-TIM1 cells were transfected with GFP or TRIM25 and either treated with 250μM of T705 (Favipiravir), or the equivalent volume of the diluent (DMSO). Relative quantification of intracellular viral RNA levels (grey) and NP-associated RNA (coloured bars) were determined as in (B). Values are presented as percentage of absolute RNA copy numbers on cells transfected with GFP. (G) Relative quantification of ZAP-associated RNA on HEK293T LacZ CRISPR-TIM1 and HEK293T TRIM25 CRISPR KO-TIM1 cells transfected with GFP (grey) or ZAP-L (red) prior to infection with EBOV trVLPs (left panel). 3 hours post-infection cells were UV cross-linked, and ZAP was immunoprecipitated from lysates. RNA extraction, cDNA synthesis and RT-qPCR analysis were performed as in (B). Values were normalized to the respective inputs and are presented relative to absolute RNA copy numbers on cells transfected with GFP. Cellular lysates and pull-down samples were analysed by western blot for HSP90, TRIM25, NP and ZAP (right panel). (H) HEK293T LacZ CRISPR, HEK293T TRIM25 CRISPR KO and HEK293T ZAP CRISPR KO cells stably expressing TIM1 were either untreated or pre-treated with 1000U/ml of IFN-I prior to infection with EBOV trVLPs. Relative quantification of intracellular viral RNA levels (grey) and NP-associated RNA (coloured bars) were determined as in (B). Values are presented as percentage of absolute RNA copy numbers on cells non-treated with IFN. (I) HEK293T LacZ CRISPR, HEK293T TRIM25 CRISPR and HEK293T ZAP CRISPR KO cells stably expressing TIM1 were pre-treated overnight with increasing concentrations of IFN-I prior to infection with EBOV nanoluciferase trVLPs. EBOV trVLP nanoluc reporter activities were measured 48 hours post-infection and data is shown as a percentage of untreated for each cell line individually. (J). Proposed model for the mechanism associated with the antiviral activities of TRIM25 and ZAP against EBOV trVLP. EBOV viral particle enters the cells by macropinocytosis, followed by NPC1-dependent fusion with the cellular endosomal membrane. Once in the cytoplasm, TRIM25 is recruited to the viral particle through an interaction with EBOV NP protein, leading to the ubiquitination of both viral target and TRIM25 itself. This results in the displacement of the viral RNA genome from the vRNP, followed by its recognition by ZAP in a way dependent of the genome’s CpG content and subsequent impact in the transcription and replication of EBOV trVLP.