Figure 1:
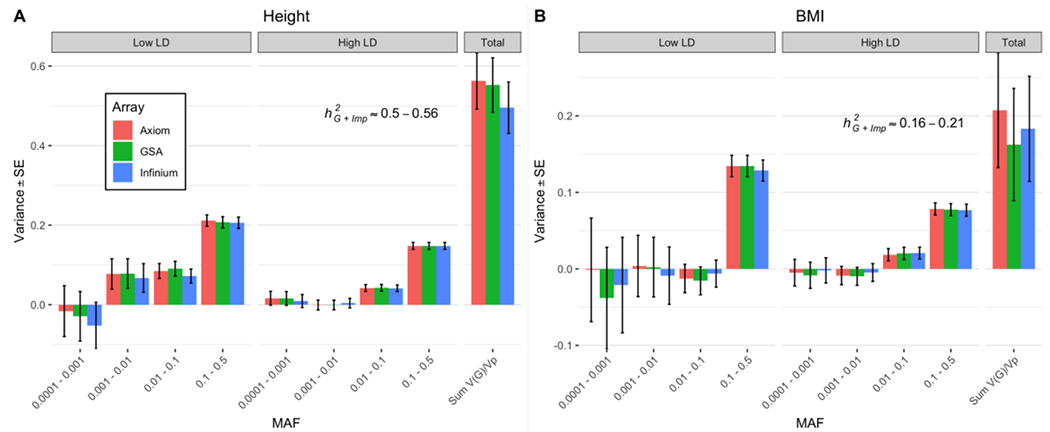
GREML-LDMS estimates with 8 bins (2 LD bins for each of the 4 MAF bins) correcting for 20 PCs (calculated from LD-pruned HM3 SNPs) after imputing SNPs from Illumina InfiniumCore24, GSA 24 and Affymetrix Axiom arrays using Haplotype Reference Consortium reference panels for N=25,465 samples. (A) Estimates of for height are between 0.50-0.56 (SE 0.06-0.07). (B) Estimates for BMI are between 0.16-0.21 (SE 0.07). The large SEs of the estimates for variants with MAF between 0.0001 to 0.001 can be explained by the large number of imputed variants in this MAF bin because the sampling variance of a SNP-based heritability estimate is proportional to the effective number of independent variants 37. Between ~19.0M and ~20.0M variants in total are included in the analysis. The number of variants in each of the 4 MAF bins (twice the number in each LD bin) can be found in Supplementary Figure 8.
