Figure 1: the OpenCell library.
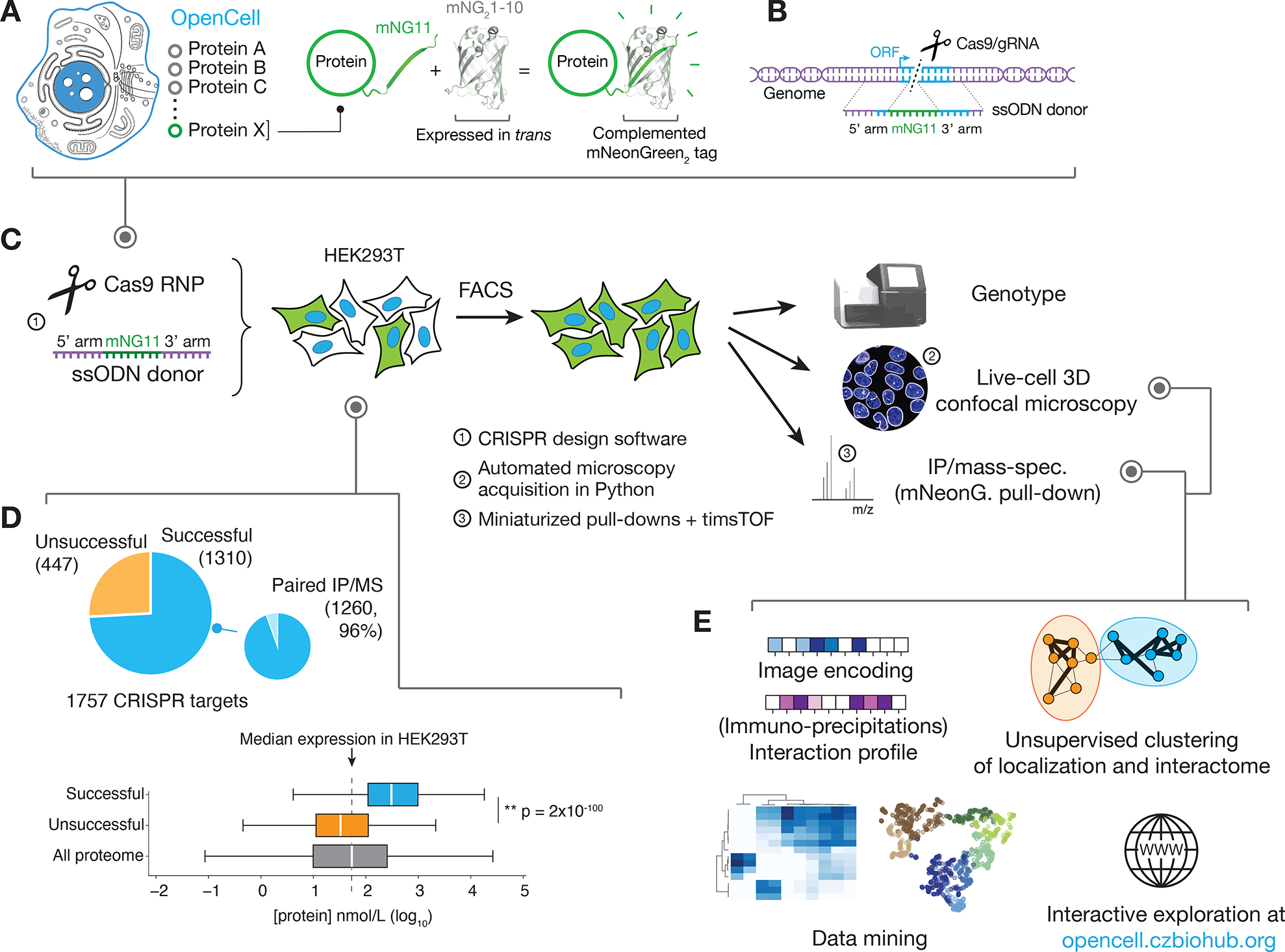
(A) Functional tagging with split-mNeonGreen2. In this system, mNeonGreen2 is separated into two fragments: a short mNG11 fragment, which is fused to a protein of interest, and a large mNG21-10 fragment, which is expressed separately in trans (that is, tagging is done in cells that have been engineered to constitutively express mNG21-10). (B) Endogenous tagging strategy: mNG11 fusion sequences are inserted directly within genomic open reading frames (ORFs) using CRISPR-Cas9 gene editing and homologous recombination with single-stranded oligonucleotides donors (ssODN). (C) The OpenCell experimental pipeline. See text for details. (D) Successful detection of fluorescence in the OpenCell library. Out of 1757 genes that were originally targeted, fluorescent signal was successfully detected for 1310 (top panel). Low protein abundance is the main obstacle to successful detection. Bottom left panel shows the full distribution of abundance for all proteins expressed in HEK293T vs. successfully or unsuccessfully detected OpenCell targets; boxes represent 25th, 50th, and 75th percentiles, and whiskers represent 1.5x interquartile range. Median is indicated by a white line. P-value: Student’s t-test. (E) The OpenCell data analysis pipeline, described in subsequent sections.
