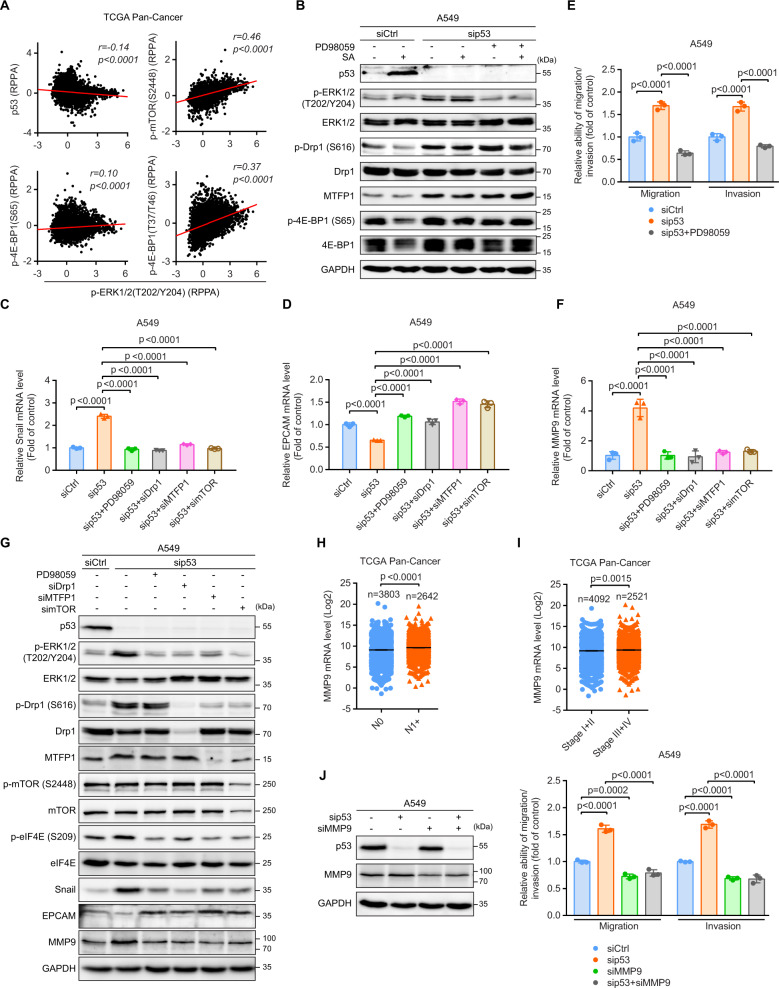
Fig. 6. Activation of mTORC1/MTFP1/Drp1/ERK1/2 signaling axis is required for the EMT switch, MMP9 elevation, and cancer dissemination upon WT p53 loss.
A Correlations between the RPPA levels of p-ERK1/2 (T202/Y204) and the indicated proteins (n = 7694 samples). Data were extracted from TCGA. B Immunoblot of the indicated proteins in siCtrl, sip53, and PD98059-treated sip53 (sip53 + PD98059) A549 cells with and without 20 µM SA treatment for 24 h. GAPDH was used as a loading control. qRT-PCR analysis of the mRNA levels of (C) Snail and (D) EPCAM in siCtrl, sip53, sip53 + PD98059, sip53 + siDrp1, sip53 + siMTFP1, and sip53+simTOR A549 cells. E Transwell assays for siCtrl, sip53, and sip53 + PD98059 A549 cells. F qRT-PCR analysis of MMP9 mRNA expression in siCtrl, sip53, sip53 + PD98059, sip53 + siDrp1, sip53 + siMTFP1, and sip53+simTOR A549 cells. G Immunoblot of the indicated proteins in siCtrl, sip53, sip53 + PD98059, sip53 + siDrp1, sip53 + siMTFP1, and sip53+simTOR A549 cells. GAPDH was used as a loading control. MMP9 mRNA expression in (H) N0 and N1 + or (I) stage I + II and III + IV tumors. Data were extracted from TCGA. J Transwell assays for siCtrl, sip53, MMP9-silenced (siMMP9), and p53/MMP9 double-knockdown (sip53 + siMMP9) A549 cells. Error bars represent mean ± SD (C–F and J) or SEM (H and I). Data were analyzed by one-way ANOVA with Tukey’s multiple comparisons test (C–F and J) or two-tailed unpaired Student’s t test (H and I).