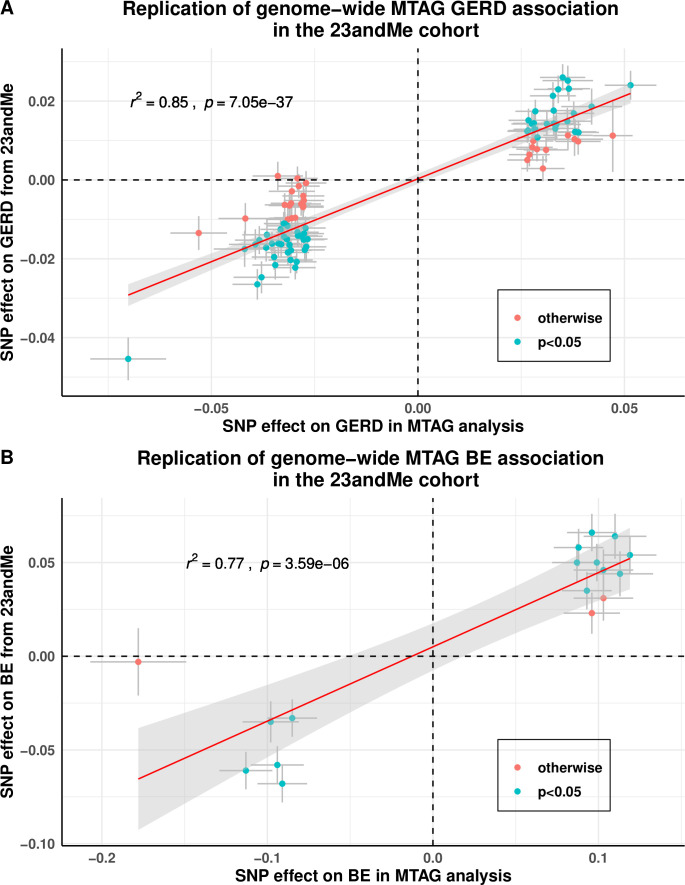
Figure 4.
Replication of GERD and BE association for the genome-wide significant loci from MTAG analysis on GERD and BE in the 23andMe cohort. A refers to the findings for GERD; (B) for BE. Data points that are shaded in blue are those that have a Bonferroni corrected p value below 0.05/88 for GERD and 0.05/17 for BE. Both the x- and y-axes represent log(OR) for GERD/BE and points are plotted with error bars representing one SE. Most of our GERD and BE loci showed strong evidence of being replicated, although the estimated effect size for GERD/BE in the 23andMe cohort were on average smaller than those estimated in the MTAG analysis (slope ~0.5 for both traits). BE, Barrett’s oesophagus; GERD, gastro-oesophageal reflux disease; MTAG, multitrait analysis of genome-wide association studies, SNP, single nucleotide polymorphism.