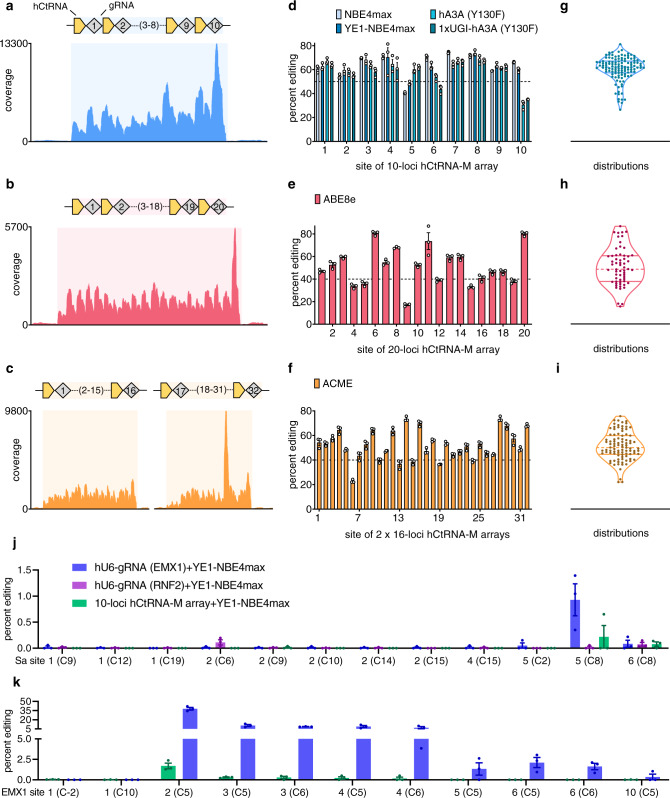
Fig. 3. DAP strategy enables large-scale MBE with minimal off-target effect.
a–c Schematic of RNA-seq results mapped onto the hCtRNA-gRNA multiplex arrays. d CBE multiplex editing of 10 loci using array shown in a with dash line indicating 50% editing. CBE architecture of each variant in d is shown in Supplementary Fig 12. e ABE8e multiplex editing of 20 loci using array shown in b with dash line indicating 40% editing. f 31-loci MBE using dual-deaminase base editor ACME and two 16-loci arrays in c, with dash line indicating 40% editing. gRNA 14 and gRNA 28 are identical and exhibit the same results in f. g Violin plot showing the value distribution of all replicates of d with solid lines representing quartiles, 57.4%, 67.3%, and dash line indicating median, 62.3%. h Violin plot showing the value distribution of all replicates in 20-loci MBE, with solid lines representing quartiles, 37.78%, 60.69%, and dash line indicating median, 48.68%. i Violin plot showing the distribution of all replicates of 31-loci MBE, with solid lines representing quartiles, 44.61%, 59.86%, and dash line indicating median, 50.12%. All tests were performed in HEK293T cells and analyzed using NGS. j Cas9-independent off-target editing evaluation of MBE at five Sa (SaCas9) sites (1,2,4,5,6) used in orthogonal R-loop assay42. k Cas9-dependent off-target editing evaluation of MBE at seven GUIDE-seq44 identified EMX1 off-target sites. Detailed protospacers, edited bases, amplicons, primers, and plasmid maps of the relevant figures are available in Supplementary Tables 1 and 5. Error bars represent mean ± s.e.m. from n = 3 replicates.