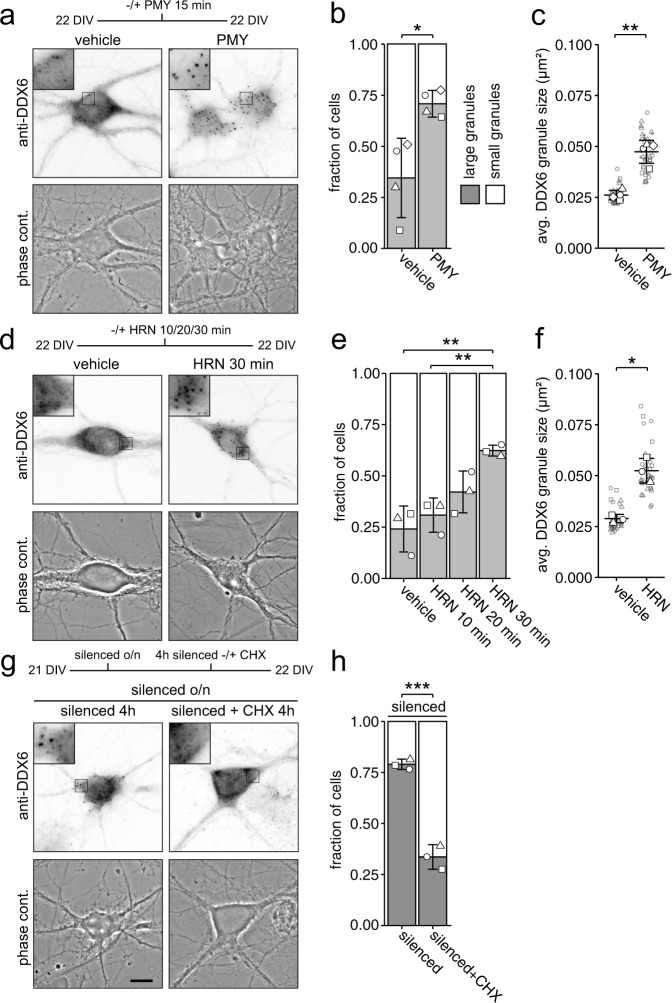
Fig. 4. DDX6 granule assembly is facilitated by the availability of cytoplasmic non-translating mRNAs.
a, d, g Experimental outline and representative examples of DDX6 immunostainings and phase contrast pictures of 22 DIV hippocampal neurons in culture under 15 min vehicle or puromycin (25 µM) treated conditions (a), under 30 min vehicle (DMSO) or harringtonine (2 µg/mL) treated conditions (d), and under silenced (100 µM CNQX, 50 µM AP5, 1 µM TTX) conditions, followed by 4 h additional silencing or silencing + CHX (g). Abbreviations: PMY = puromycin, HRN = harringtonine, CHX = cycloheximide. Boxed regions in images are displayed as magnified insets. Scale bars 10 µm. b, e, h Bar plots displaying quantification of cell population by fraction of cells containing either large or small DDX6 granules as exemplified in (a), (d) and (g). Distinct dot symbols indicate biological replicates. At least 100 cells/condition/experiment were quantified. n = 4 (b) and n = 3 (e, h) biologically independent experiments). c, f Dot plots displaying average DDX6 granule size of individual cell bodies. Small gray symbols represent single cells while larger white symbols indicate the average of each replicate. Data represents mean of three or four independent neuronal cultures. n = 4 (c) and n = 3 (f) biologically independent experiments). Asterisks represent p-values obtained by two-sided Student’s t-test (b, c, f, h) or Tukey’s test post-hoc to one-way ANOVA analysis (e) (*p < 0.05, **p < 0.01, ***p < 0.001). p = 0.0276 (b), p = 0.0022 (c), F3,8 = 0.00333 (e), p = 0.0096 (f), p = 0.00084 (h).