Figure 3.
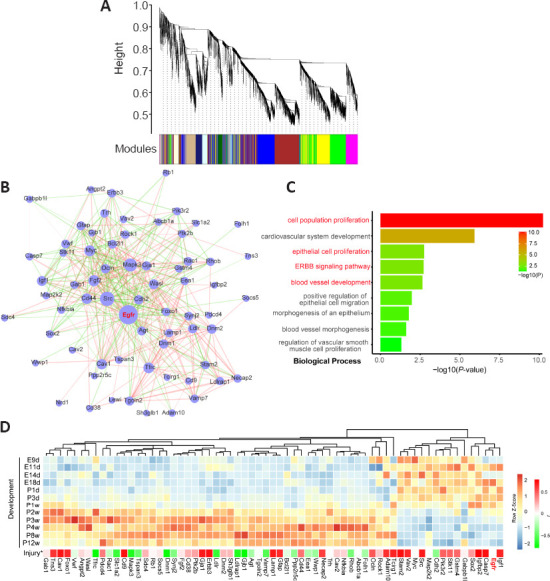
A co-expression module with Egfr inferred by weighted gene co-expression network analysis (WGCNA) based on an RNA-seq dataset of rat spinal cord development.
(A) Co-expressed modules inferred by the WGCNA method based on an RNA-seq dataset of rat spinal cord development and a blue module containing Egfr. (B) The subnetwork of first neighbors of Egfr and protein-protein interactions (PPIs) predicted from genes in the blue module. PPIs in the blue module were retrieved from the STRING database. (C) Gene ontology analysis of biological processes shows enrichment of genes in the subnetwork of first neighbors of Egfr. (D) Gene expression profile of genes in the subnetwork of first neighbors of Egfr. * Indicates the Pearson’s correlation (r) calculated based on the spinal cord injury dataset from Yu et al. (2019).
