Fig. 3.
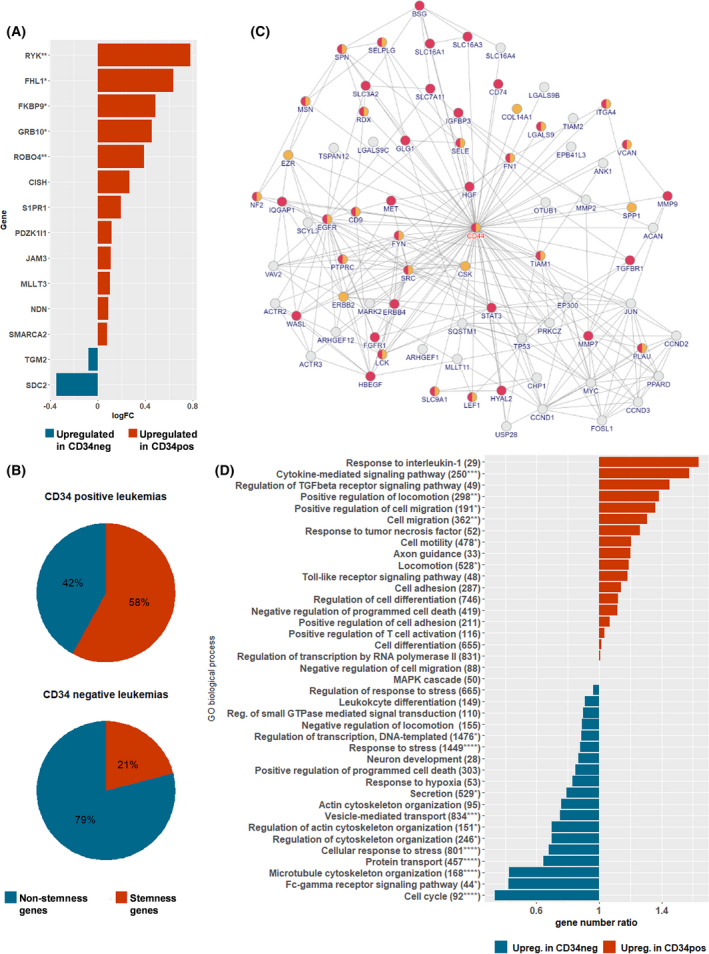
Annotations of differentially expressed genes between CD34‐positive and CD34‐negative leukemias. (A) Fourteen genes, found to be upregulated in normal, quiescent HSCs and validated by qPCR by Forsberg et al. [41] were evaluated for expression in CD34‐positive (n = 87) and CD34‐negative leukemias (n = 18). Expression of 12/14 was higher in CD34‐positive leukemias, two (ROBO4 and RYK) significantly after FDR correction (Benjamini–Hochberg). *P < 0.05, **q < 0.05. (B) The top 50 differentially expressed genes between CD34‐positive and CD34‐negative leukemias were evaluated for association with stemness in the literature (Table S3). 18/26 for CD34 positive vs 8/24 for CD34 negative were found to associate with stemness, P = 0.0225, Fisher's exact test. (C) Network 5 with seed protein CD44. Edges represent known protein interactions, and nodes represent proteins, with gene names encoding the proteins in capital letters below. Yellow nodes mark genes involved in cell adhesion (cell adhesion GO:0007155, regulation of cell adhesion GO:0030155, positive regulation of cell adhesion GO:0045785, and negative regulation of cell adhesion GO:0007162), red nodes mark cell migration (cell migration GO:0016477, regulation of cell migration GO:0030334, positive regulation of cell migration GO:0030335, and negative regulation of cell migration GO:0030336). (D) Direction of gene expression in enriched GO biological processes from PPI networks. Processes, found to be overrepresented in at least one of the 10 PPI networks (Table S4) and comprising more than 10 genes, are included. For each GO process, the number of genes upregulated more than 0.1 log2FC in CD34‐positive vs CD34‐negative leukemias and vice versa were registered and the ratio between the two illustrated. The name of the process is followed by the number of genes in the process and a P‐value testing distribution of genes upregulated in CD34 positive vs CD34 negative within each GO process vs distribution in remaining genes (total: 8709 upreg. in CD34 negative, 8490 upreg. in CD34 positive) using Chi‐square test with Yates' correction (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
