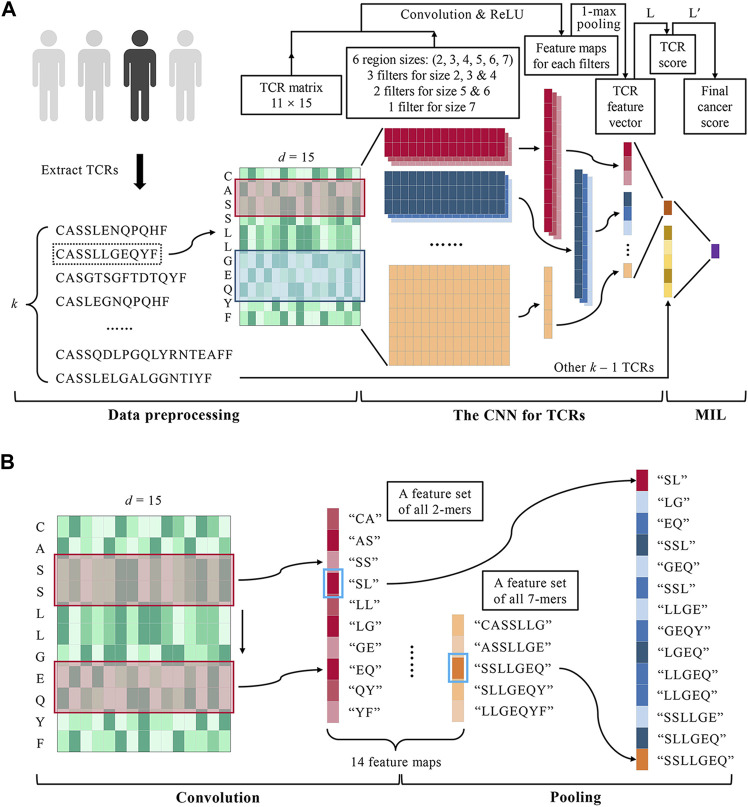
FIGURE 1.
DeepLION for accurate TCR repertoire prediction. (A) The workflow of DeepLION is divided into three parts: data preprocessing, the CNN for TCRs, and MIL. During data preprocessing, the top k most abundant TCR sequences were extracted from each repertoire after removing unqualified sequences and they were encoded into matrixes by the Beshnova matrix. The CNN for TCRs consisted of 14 convolution filters covering six various region sizes, 1-max pooling operations, and a one-layer linear classifier L. The TCR matrixes were input to the CNN and their scores were output. In the MIL part, DeepLION employed another one-layer linear classifier L′ to aggregate k TCR scores to predict the repertoire. (B) The details of the convolution and pooling operations of CNN in DeepLION. When a 2 × d convolution filter (the red box) performed a complete convolution operation on the TCR matrix from top to bottom, it could be regarded as extracting the biochemical features of the 2-mers such as "CA", "AS", etc., and then a 10 × 1 feature map, a feature set of all 2-mers, was generated. Other filters performed similar convolution operations and 14 feature maps were obtained. The maximum value of each map (marked with a blue box) was selected by a 1-max pooling operation, which could be viewed as the feature of the z-mers most likely to be the cancer-specific motif. These features were interconnected to generate a 14 × 1 TCR feature vector.