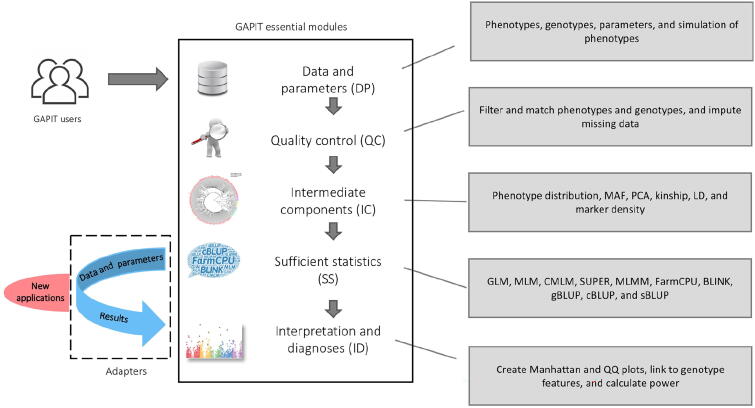
Fig. 1.
GAPIT essential modules and adapters to external packages. GAPIT version 3 was designed to have five sequential modules and multiple adapters that connect external software packages. The first module (DP) is responsible to process input data and parameters from users. The second module (QC) is responsible for quality control, including missing genotype imputation. The third module (IC) provides intermediate results, including MAF, PCA, kinship, LD analysis, and maker density distribution. The fourth module (SS) contains multiple adapters that convert input data into sufficient statistics, including maker effects, P values, and predicted phenotypes. The current adapters include GLM, MLM, CMLM, SUPER, MLMM, FarmCPU, BLINK, gBLUP, cBLUP, and sBLUP. The fifth module (ID) provides the interpretation and diagnosis on the final results, including P values illustrated as Manhattan plots and QQ plots. GAPIT, genomic association and prediction integrated tool; DP, data and parameters; QC, quality control; IC, intermediate components; MAF, minor allele frequency; PCA, principal component analysis; LD, linkage disequilibrium; GLM, general linear model; MLM, mixed linear model; CMLM, compressed MLM; SUPER, settlement of MLM under progressively exclusive relationship; MLMM, multiple loci mixed model; FarmCPU, fixed and random model circulating probability unification; BLINK, Bayesian-information and LD iteratively nested keyway; gBLUP, genomic best linear unbiased prediction; cBLUP, compressed BLUP; sBLUP, SUPER BLUP; QQ, quantile–quantile.