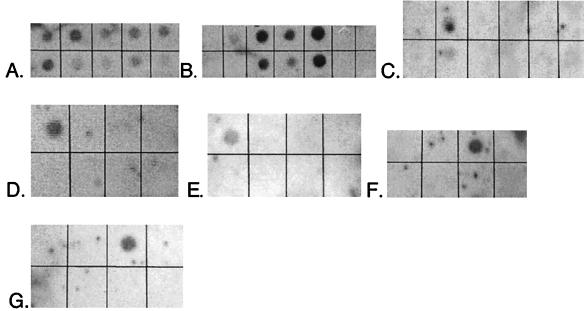
FIG. 3.
Probe specificity tests. Membranes were hybridized with the indicated probe (target group). DNAs (1 μg per spot except in bottom rows of panels B and C, where 500 ng per spot was used) are listed as they appear on membranes from top left to bottom right. (A) EUB-338 (general eubacteria). DNAs: E. coli, D. vulgaris, T. aquaticus, marsh sample, B. subtilis, D. postgatei, D. autotrophicum, D. multivorans, D. desulfuricans. (B) SRB-385 (general SRB). DNAs: E. coli, D. proprionicus, D. desulfuricans, D. postgatei, D. multivorans, B. subtilis, T. aquaticus. (Bottom row shows lower concentrations of the same nucleic acids.) (C) SRB-129 (Desulfobacter spp.). DNAs: D. autotrophicum, D. postgatei, D. vulgaris, D. multivorans, D. proprionicus, E. coli. (Bottom row shows lower concentrations of the same nucleic acids.) (D) SRB-687 (Desulfovibrio spp.). DNAs: D. vulgaris, D. multivorans, D. postgatei, E. coli, D. proprionicus, D. autotrophicum. (E) SRB-221 (Desulfobacterium spp.). DNAs: D. autotrophicum, D. desulfuricans, D. proprionicus, D. multivorans, D. postgatei, E. coli. (F) SRB-660 (Desulfobulbus spp.). DNAs: E. coli, D. multivorans, D. proprionicus, D. vulgaris, D. autotrophicum, D. postgatei, B. subtilis, D. desulfuricans. (G) SRB-814 (Desulfococcus spp.). DNAs: D. vulgaris, D. multivorans, D. postgatei, E. coli, D. proprionicus, D. autotrophicum. Images were prepared with NIH Image and Adobe Photoshop 5.0.