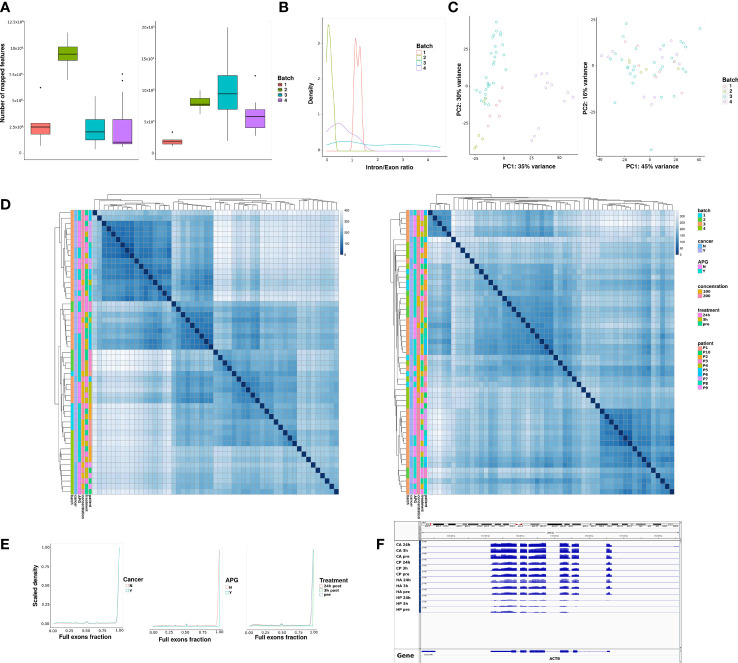
Figure 1.
Characteristics of the sequencing runs. Samples were run in four batches under different conditions. (A) Number of sequencing reads mapping to features of interest (exons) per sequencing batch, non-corrected (left), after mitochondrial transcript removal (right). (B) Intron to exon ratio per sequencing batch. (C) Principal component analysis after normalization and variance-stabilizing transformation of samples before (left) and after batch correction (right). (D) Distance matrices between samples before (left) and after (right) batch correction. Each row and column represent a single sample, and the diagonal represents samples that are identical. A higher distance score represents higher dissimilarity between a pair of samples. Sample annotation is shown by multiple variables, such as: batch, whether a patient has cancer or not, whether they received 100mg or 200mg of APG-157 or Placebo, treatment time point and patient ID. We have included the same batch and cross-batch technical replicates for a portion of the samples. (E) The distribution of exon occupancy across all genes with 2 or more exons; a value of 1 means full occupancy; a value between 0 and 1 means partial exon occupancy; genes with no exon occupancy have been removed from the dataset. (F) An IGV screenshot showing the coverage of ACTB as a representative gene with full exon occupancy.