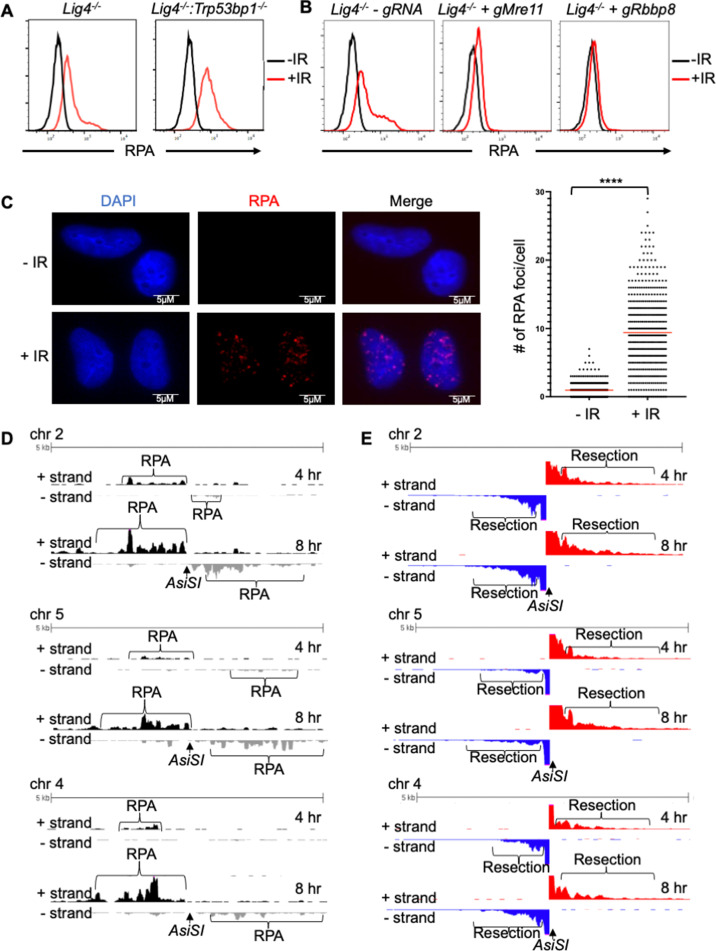
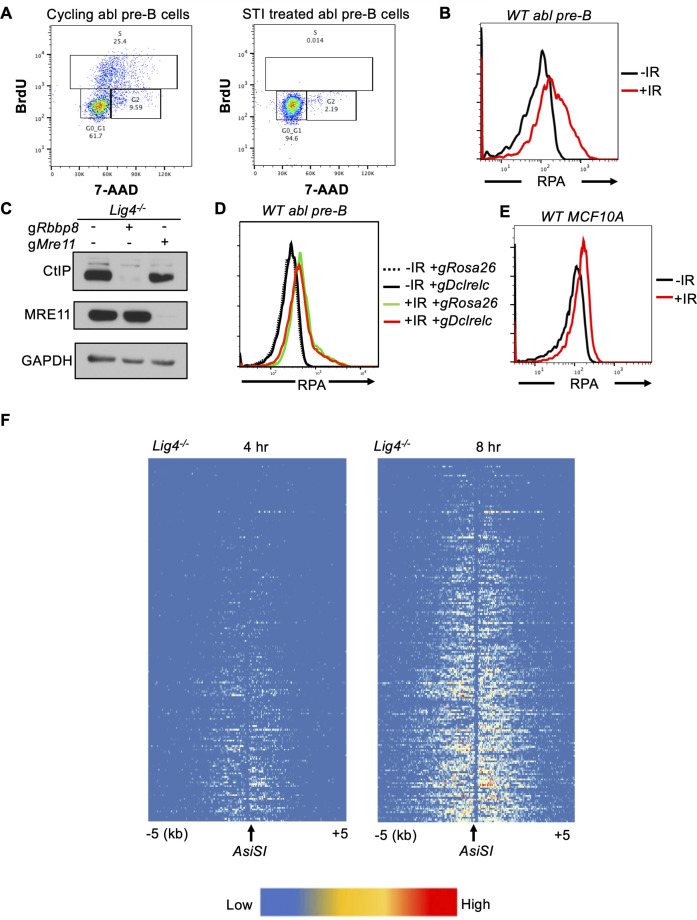
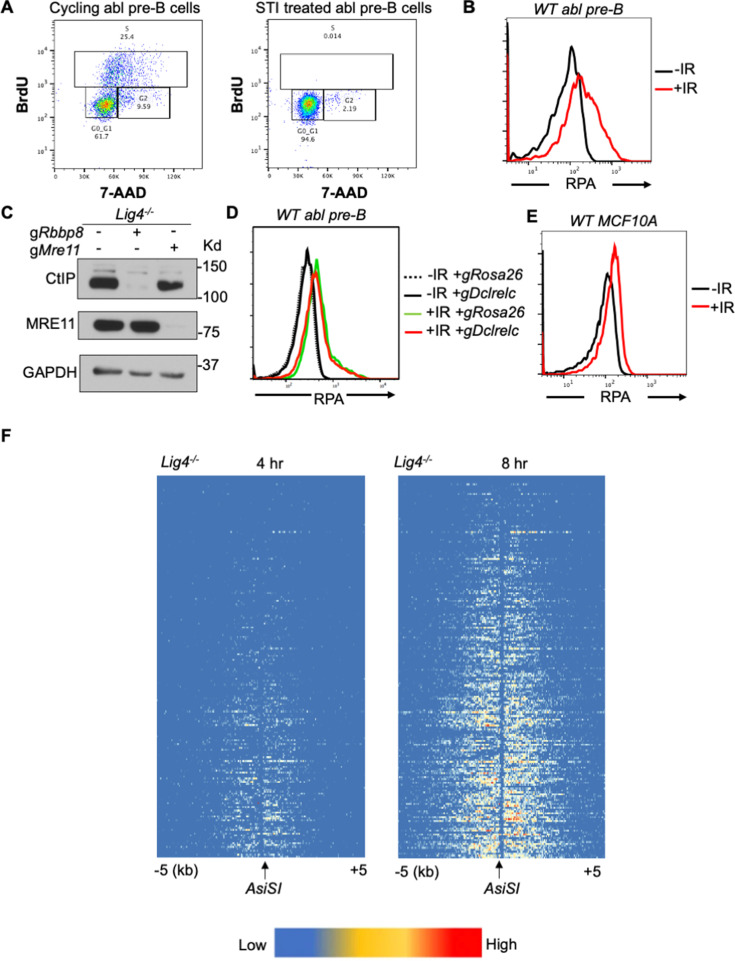
Figure 1. RPA is loaded onto ssDNA after DSBs in G0 mammalian cells.
(A) Flow cytometric analysis of chromatin-bound RPA in G0-arrested Lig4-/- and Lig4-/-:Trp53bp1-/- abl pre-B cells before and 3 hr after 20 Gray IR. Representative of three independent experiments. (B) Flow cytometric analysis of chromatin-bound RPA before and 2 hr after 15 Gy IR in G0-arrested Lig4-/- abl pre-B cells (left), Lig4-/- cells depleted of MRE11 (middle), and Lig4-/- cells depleted of CtIP (right). Representative of three independent experiments. (C) Representative images and quantification of IR-induced RPA foci from three independent experiments in G0-arrested MCF10A cells before and 3 hr after 10 Gray IR. n=365 cells in -IR and n=433 cells in +IR. Red bars indicate average number of RPA foci in - IR = 0.96 and average number of RPA foci in +IR = 9.4 (****p<0.0001, unpaired t test). (D) RPA ChIP-seq tracks at AsiSI DSBs on chromosome 2, 5, and 4 at 4 hr (top) and 8 hr (bottom) after AsiSI endonuclease induction in G0-arrested Lig4-/- abl pre-B cells. (E) Representative END-Seq tracks showing resection at AsiSI DSBs at chromosome 2, 5, and 4 at 4 hr (top) and 8 hr (bottom) after AsiSI induction in G0-arrested Lig4-/- abl pre-B cells. END-seq data is representative from two independent experiments.