Abstract
Cas12a is an RNA-guided endonuclease that is emerging as a powerful genome-editing tool. Here, we selected a target site on bacteriophage λ-DNA and used optical tweezers combined with fluorescence to provide mechanistic insight into wild type Cas12a and three engineered variants, where the specific dsDNA and the unspecific ssDNA cleavage are dissociated (M1 and M2) and a third one which nicks the target DNA (M3). At low forces wtCas12a and the variants display two main off-target binding sites, while on stretched dsDNA at higher forces numerous binding events appear driven by the mechanical distortion of the DNA and partial matches to the crRNA. The multiple binding events onto dsDNA at high tension do not lead to cleavage, which is observed on the target site at low forces when the DNA is flexible. In addition, activity assays also show that the preferential off-target sites for this crRNA are not cleaved by wtCas12a, indicating that λ-DNA is only severed at the target site. Our single molecule data indicate that the Cas12a scaffold presents singular mechanical properties, which could be used to generate new endonucleases with biomedical and biotechnological applications.
INTRODUCTION
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) that associate with CRISPR associated (Cas) proteins, constitute an adaptive immune system in bacteria and archaea against foreign mobile genetic elements (MGEs), such as plasmids and phages. These RNA-guided nucleases have been repurposed as versatile genome editing tools, triggering a technological revolution in the life sciences (1–3). CRISPR-Cas immunity maintains a genetic record of encounters with invading MGEs in the CRISPR array in the form of unique DNA segments (spacers), which are intercalated between short repetitive sequences (repeats). Transcription of the CRISPR array yields a long pre-CRISPR RNA (pre-crRNA), which after maturation into shorter crRNA molecules, forms a ribonucleoprotein (RNP) complex with the effector protein, resulting in a functional RNA-guided endonuclease responsible for interference. To maintain the integrity of the CRISPR array, the target DNA cleavage depends on the recognition of a short protospacer adjacent motif (PAM) sequence. The PAM sequence recognized by the effector complex is present in the invading DNA molecules but not in the CRISPR array, therefore protecting this DNA region from cleavage (4).
Depending on the type of RNP effector nucleases, CRISPR-Cas systems can be categorized into two classes (1 and 2), which are further subdivided into six types (types I through VI). Class 2 systems form an RNP complex between the multidomain effector Cas protein and a CRISPR RNA (crRNA) that contains the necessary information to target a specific nucleic acid sequence (2,5,6). Cas9 is the best characterized member of Class 2 (Type II) and it has been developed into a genome editing tool (1,3,7–11). Diverse Cas9 nuclease variants are involved in clinical trials to develop new therapies against human diseases (12). Despite its efficacy and versatility, the use of Cas9 in genome editing is limited due to toxic and possible mutagenic effects (13,14). These problems have boosted the search of CRISPR-Cas systems that can be developed into new genome editing tools. Recently, other members of Class 2 type V (Cas12a, Cas12b, CasΦ, Cas12g, Cas14) have been studied (15–22). However, they have not been as extensively characterized as Cas9.
Here we focus on Cas12a, this RNA-guided nuclease adopts a bilobed architecture with a recognition lobe (REC) and a nuclease lobe (NUC). In contrast with Cas9, which generates a blunt-end DNA cleavage next to the PAM, Cas12a introduces a 6–7-nt staggered cleavage 70 Å away from the PAM sequence using a single catalytic site in a cleft between the RuvC and NuC domains (10,23). The DNA overhang is produced by the different paths followed by the non-target (NT) and the target (T) strands to reach the catalytic pocket (24). The absence of a HNH domain in Cas12a implies that the catalytic site in RuvC cleaves both the NT- and T-strands of the target DNA. However, while the NT-strand can reach the catalytic pocket with a 5′-3′ polarity, a conformational change in the distal side of the REC and NUC lobes of Cas12a is needed to accommodate the T-strand with the proper polarity for cleavage. This results in the faster severing of the NT- compared to the T-strand (25,26). In addition, an unspecific double-stranded DNase activity has been reported (27) and an indiscriminate catalytic single-strand (ss) DNase activity was also discovered in Cas12a. The ssDNA secondary activity is triggered by binding of the target DNA or ssDNA complementary to the crRNA, thus mimicking the T-strand (28). Therefore, Cas12a is capable of precise cleavage of target DNA and indiscriminate degradation of ssDNA after activation. This activity has further expanded the possible applications of Cas12a to molecular diagnostics (28).
Cas12a is an attractive candidate for genome editing, as it seems to display higher precision than Cas9 (29) and it presents room for further improvement (8,9). In addition, Cas12a can alleviate the size problem in delivery via viral vectors due to its small crRNA (∼40nt) (30–34), and its unique RNase activity, which enables Cas12a to process its pre-crRNA into mature crRNAs (10,35). This activity of Cas12a has been exploited for use in multiplex gene regulation and has been shown to successfully edit multiple endogenous targets simultaneously through delivery of multiple crRNAs on a single plasmid (30,36).
To decipher the singularities of the Cas12a RNA-guided endonuclease, we selected a target site on λ-DNA and analyzed its mechanical properties, combining optical tweezers with confocal microscopy to dissect its binding and phosphodiester hydrolysis on force-stretched DNA in real time. Our results show that wild type (wt) Cas12a binding is more promiscuous at high forces, and it cleaves the selected target when low forces are applied to the λ-DNA. Structure-function studies of Cas12a provided snapshots of the molecular details of Cas12a (24–26,37,38). Three regions, termed the ‘linker’, the ‘lid’ and the ‘finger’, build a network of checkpoints along the main cavity of the protein that monitor the assembly of the hybrid between the crRNA and the T-strand to open the catalytic site in the RuvC/Nuc pocket of Cas12a (25). Based on the structural data we performed mutagenesis in the linker and the lid, as mutations in the finger rendered insoluble versions of Cas12a (25). We generated several variants and three of them displayed different cleavage properties in bulk experiments (25). The Cas12a_M1 variant contains mutations in the linker and only cuts ssDNA indiscriminately after activation with an oligonucleotide complementary to the crRNA guide (25). The Cas12a_M2 and Cas12a_M3 variants display double mutations in the lid (25) (Materials and Methods). Cas12a_M2 conserves the specific dsDNA cleavage but does not unleash indiscriminate ssDNA degradation, while Cas12a_M3 generates nicks on the dsDNA target. These variants are potential candidates for biomedical and diagnostic applications.
MATERIALS AND METHODS
Plasmid preparation
All experiments in this manuscript were performed with the Francisella novicida (Fn) Cas12a gene. All cloning and site directed mutagenesis were performed as previously described in (25). Variants of Cas12a were generated based on information from structural studies performed in (25). The mutations in the variants are listed as follows: Cas12a_M1 contains a substitution of the key residues in the protein region the REC-linker from L324 to K335 (LFKQILSDTESK) to 324-GGGAGASAGGSG-335. Cas12a_M2 contains the mutations Q1025G and E10128G. Cas12a_M3 contains the mutations K1013G and R1014G. Cas12a_KO contains mutations in the active site, D917A and E1006A. All genes were cloned into the pET-21a vector.
Protein expression and purification
The target construct containing pET21-FnCas12a was transformed into Escherichia coli BL21 star (DE3) cells carrying the pRare2 plasmid (that provides seven tRNAs recognizing rare codons). A single colony containing both plasmids was selected to inoculate a culture of 50ml LB media, with 50 μg/ml ampicillin and 25 μg/ml chloramphenicol, grown overnight at 37°C, which was then used to inoculate 1L fresh LB media containing the same quantities of antibiotics and grown at 37°C until an OD600 between 0.7 and 0.9 was reached. The large-scale culture was then induced with 0.5mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and grown at 37°C for an additional 3 h. The cells were then harvested, resuspended in lysis buffer [50 mM HEPES pH 7.5, 2M NaCl, 5 mM MgCl2, 0.5 mM TCEP, 1 tablet of complete Inhibitor cocktail EDTA free (Roche), per 50 ml, 5 U/ml Benzonase, 160 μg/ml lysozyme], and lysed by sonication for 10 min with 15 s ON and 20 s OFF cycle. The lysate was then centrifuged and filtered to remove cell debris and insoluble particles, and then loaded onto a 5 ml crude HisTrap™ FF column (Cytiva) equilibrated in 50 mM HEPES pH 7.5, 500 mM NaCl, 10 mM Imidazole, 0.5 mM TCEP. The protein was eluted through a step gradient of 3%, 8% and 50% of the following buffer [50 mM HEPES pH 7.5, 500 mM NaCl, 500 mM Imidazole, 0.5 mM TCEP]. Enriched protein fractions from the 50% elution were loaded onto a 5 ml HiTrap Heparin HP column (Cytiva) equilibrated with 50 mM HEPES pH 7.5, 100 mM NaCl, 0.5 mM TCEP. Elution was performed through a linear gradient of 0–100% of the following buffer [50 mM HEPES pH 7.5, 1 M NaCl, 0.5 mM TCEP]. Enriched protein fractions from the elution were concentrated and loaded onto a HiLoad Superdex 200 16/60 column (Cytiva) equilibrated in 50 mM Bicine pH 8.0, 150 mM KCl, 0.5 mM TCEP. Protein rich fractions were concentrated using 50 kDa Amicon® Ultra-15 Centrifugal Filter Units, flash frozen in liquid nitrogen and stored at −80°C. All the variants were also expressed and purified using the above protocol.
crRNA design for single molecule experiments
The region 29.711 to 29.732 kb on the Enterobacteria phage λ DNA was chosen as the prospective target because it contained the FnCas12a PAM sequence (TTN). Five different crRNAs (crRNA-1 to 5) at different locations on the λ-DNA were designed and tested for cleavage (Supplementary Figure S1 and Table S1). A target region (spacer) of 22nt was placed on the 3′ end of the repeat sequence 5′-AAUUUCUACUGUUGUAGAU-3′ to form a 41nt long crRNA. We used a 22nt spacer to ensure the Cy5 label did not interfere with binding to the target. The best activity was observed for crRNA-1, which was used in the single molecule experiments. This crRNA was purchased from IDT, with fluorescent labels on the 3′ end (Cy5), 3′-Cy5-labeled crRNA-1.
Bulk cleavage assays
The purified protein was incubated with the crRNA-6 (rArArU rUrUrC rUrArCr UrGrUr UrGrUr ArGrAr UrGrAr GrArA rGrUr CrArU rUrUrA rArUrA rArGrGr CrCrArCrU) in the molar ratio of protein:RNA 1:1.3, at room temperature for 15mins to form the ribonucleoprotein complex (RNP) in reconstitution buffer A with the following composition: 20 mM Bicine pH 8.0, 150 mM KCl, 5 mM MgCl2. The fluorescently labelled dsDNA target was prepared by annealing the labelled target TS-6 (CGA TGC ATG CAG TGG CCT TAT TAA ATG ACT TCT CTA ACG/36-FAM/) and non-target strands NTS-6 (/56-FAM/CGT TAG AGAAGT CAT TTA ATA AGG CCA CTG CAT GCA TCG) according to the manufacturer's instructions (both oligos were independently purchased from IDT). For bulk cleavage assays with crRNA-1, TS-1-long and NTS-1-long were annealed to make the dsDNA substrate (see Supplementary Table S1). The RNP from the former reaction was then incubated with the DNA substrate in a molar ratio of RNP:DNA 1:1.7, at 37°C for 30 min, and then were stopped after incubation by adding equal volumes of a stop solution (8 M urea and 100 mM EDTA pH 8.0), followed by incubation at 95°C for 10 minutes. For the bulk cleavage assays of Cas12a indiscriminate ssDNase activity, the RNP is first activated using an activator ssDNA molecule, either λ-activator (See Supplementary Table S1) or Activator TS (See Supplementary Table S1), and then incubated with an ssDNA substrate Unspecific ssDNA (see Supplementary Table S1), and then stopped using the above stop solution. The stopped reactions were then loaded onto 15% Novex TBE–urea gels (Invitrogen) and run according to the manufacturer's instructions and imaged using an Odyssey FC Imaging System (Li-Cor).
Ratio of cleavage of the target and non-target strands
The RNP was prepared as described as above in the bulk binding assays, and then incubated with the annealed dsDNA substrate, also described above, in the same molar ratio. The reaction was monitored at several time points at 1, 5, 10, 30, 60, 90, 120 and 180 min by taking the reaction at those time points and stopping it with equal volumes of stop solution as described above, followed by incubation at 95°C for 10 min. The samples (containing 4 pmol of dsDNA substrate) were then loaded onto 15% Novex TBE–urea gels (Invitrogen) and run according to the manufacturer's instructions and imaged using an Odyssey FC Imaging System (Li-Cor). The intensity (I) of the DNA bands was quantified using ImageStudio. Considering the (I)t = 0 = 4 pmol, the formation of products at different time points was calculated using the following formula pmolest = n = 4 pmol * (I)t = n/(I)t = 0, where n is the different time points. The average of three independent experiments was used in the plot.
Complex formation for single-molecule experiments
For optical tweezers experiments, Cas12a protein was complexed 1:1 with 3′-Cy5-labeled crRNA-1 (rArArUr UrUrCr UrArCr UrGrUr UrGrUr ArGrAr UrUrGr UrArA rArGrA rArArC rArGrUr ArArG rArUrA rArU/3Cy5Sp/) in a final concentration of 5 uM and low volume of 20 ul in buffer A (20 mM Bicine pH 8.0, 150 mM KCl, 5 mM MgCl2). After adding the Cas12a protein and crRNA-1 together, the sample was left at RT for 15 min after which it was kept on ice until further processing. For the experiments including the activator oligo, the Cas12a–crRNA complex was incubated with λ-activator, a 40nt activator ssDNA (CAT CGG GTT GAG TAT TAT CTT ACT GTTT CTT TAC ATA AAC) at a ratio 1:2, 10 uM final concentration, for 15–30 min at 25ºC, after which the sample was kept on ice. For optical tweezers experiments, the 5 uM complex stock solution was diluted in either buffer A (20 mM Bicine pH 8.0, 150 mM KCl, 5 mM MgCl2) or buffer B (20 mM Bicine pH 8.0, 150 mM KCl), depending on whether the complex would be used for cleavage or binding measurements respectively, to a working concentration of 1nM (1:5000).
Optical tweezers assay
Single-molecule experiments were performed at room temperature using a correlative confocal fluorescence - optical tweezers microscope (LUMICKS, C-Trap). Two traps were generated by a 1064 nm laser passing through a 60× Water Immersion (NA 1.2) objective. One trap was beam-steered by controlling a piezo mirror, while the other trap provided force measurements. Force measurements were performed by back-focal plane interferometry using a position-sensitive detector. The combination of micro-stage movements with a 5-channel microfluidics system allowed rapid in situ construction and examination of dumbbell assays.
Surface passivation
To ensure a stable concentration, the channels harbouring Cas12a–crRNA complex (‘protein channel’ hereafter) were passivated in multiple steps. After completion of the cleaning protocol, the protein channels were incubated with 0.1% BSA (in MQ) for 20 min under flow. Then, the channels were incubated for 20 min in 0.5% Pluronics (in MQ), again under flow. Finally, at least 250 ul of the [1 nM] complex working solution was loaded in the protein channels and left in the system overnight. The next day, the complex working solution was replaced with a fresh working solution before starting experiments.
Dilution and oxygen scavengers
On the day of experiments, a fresh working solution was made from the complex stock solution that was prepared the day before by diluting either in Buffer A (for cleavage experiments) or in Buffer B (for binding experiments and wildtype cleavage control with no Mg2+ ions) to a working solution of 1 nM (1:5000). This working solution was supplemented with fresh Oxygen Scavengers (0.65% Glucose (d-(+)-glucose, Sigma) (39); 165U/ml glucose oxidase (glucose oxidase from Aspergillus niger, Sigma) (2); 2170 U/ml catalase (catalase from bovine liver, Sigma) (40). The final working solution was replaced with fresh buffer every 90 min.
DNA templates and beads
As a template, λ-DNA (48.5 kb) was used containing biotin labels (LUMICKS, Amsterdam). For experiments using dsDNA templates, λ-DNA was labelled with biotin labels on complementary strands. For experiments using ssDNA, λ-DNA contained biotin labels on either side of the same strand, allowing the other strand to be melted off at higher tension (41). DNA molecules were captured between two trapped streptavidin-coated polystyrene beads (Ø 4.38 μm, Spherotech).
Data acquisition
Workflow binding assays
Beads were optically trapped and calibrated using the power spectrum method (42). Next, DNA was tethered between the two beads by use of the multichannel laminar flow cell. The presence of a single DNA molecule was confirmed by comparing the force-distance curves to the inherent mechanical force-extension expected for single dsDNA (43). In the case of ssDNA experiments, dsDNA molecules were stretched to high forces, causing the DNA to melt (41). For binding assays, the construct was subsequently moved to the protein channel. The applied force on the DNA molecule was increased in a stepwise manner by increasing the distance between the beads, maintaining at least 120sec at each force level. For all binding assays (all mutants), the experiments were performed in the absence of Mg2+.
Workflow cleaving assays
Constructs were generated as described above. All experiments were performed in the presence of Mg2+ ions, except for the wild type control with no Mg2+. For static cleavage experiments, constructs were moved into the protein channel under a constant force of 20 pN and kept there for at least 200 s. For dynamic cleavage experiments, constructs were moved into the protein channel at low force, after which a force-extension curve was initiated and repeated for at least 4 times (taking ±200 s) or until cleavage occurred. Cleavage is defined as a loss of tether, marked by a sudden decrease in force. (Figures 1C and 4B).
Figure 1.
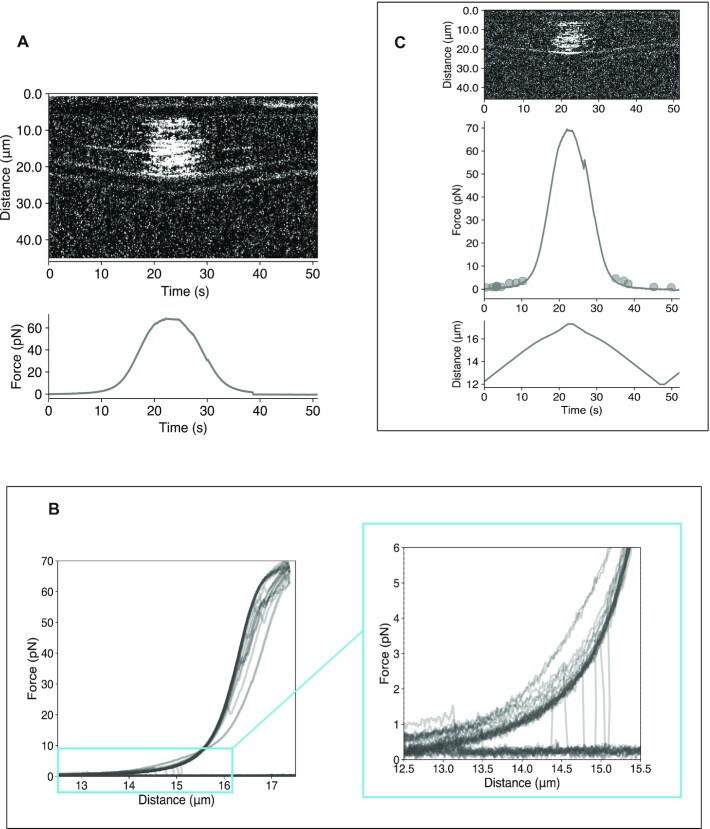
Experimental setup of Cas12a binding and cleavage onto λ-DNA. (A) Schematic of the experimental assay (left) and the microfluidic chip (right). The RNP assembly of the target DNA includes the NT-strand (green), T-strand (grey), crRNA (red) and the location of the fluorescent probe. (B) Example of kymograph showing Cas12a binding (red channel) at 10 pN. A single-pixel line scan is made along the DNA axis and plotted over time. Intensity peaks indicate binding events. (C) Representative force-distance curves in the presence and absence of Mg2+. In the absence of Mg2+, the force increases with increasing inter-bead distance, while in the presence of Mg2+, the DNA tether ruptures at low forces, as marked by a sudden drop in force. The forward curves are black with some transparency and the backwards curves are red. Hysteresis is observed in some of the reverse curves.
Figure 4.
DSB occurs exclusively at low forces in the presence of Mg2+. (A) Kymograph showing Cas12a binding during a force–distance cycle. The increment of inter-bead distance generates a increase in force applied to the tether. After relaxation, a DSB can be observed by the sudden drop in force. The right panel 2D scan shows Cas12a bound to the ends of the flow-extended DNA tether after DSB. (B) Force-extensions curve of DNA tethers in the presence of Cas12a and Mg2+ (n = 15). The inset depicts the sudden drop in tension indicative of a DSB generation. (C) Summary of all DSB events measured in the presence of Cas12a and Mg2+ during force–distance cycles (n = 14/15). The top panel shows a representative kymograph of Cas12a binding during force increase. In the middle panel, all DSB events (n = 14, points) are plotted on a force-distance curve from the corresponding kymograph. All 14 DSB events happened at forces ≤5 pN. Nine of them occurred on the force ramp up, while 5 were observed on the force ramp down. The bottom panel depicts the distance between the two beads versus time.
Fluorescence imaging
Fluorescence microscopy was achieved by imaging the construct with a confocal system using 532 nm (for bead visualization) and 638nm excitation (for Cy5) lasers and corresponding avalanche photodiodes (APDs). Confocal pixel size was set to 100 nm and the pixel dwell time was 0.05 ms. For kymographs, a single pixel height line scan along the DNA axis was scanned and plotted over time (y = DNA molecule, x = time).
Data analysis
120s binding profile (binding location)
To analyse binding profiles (‘profiles’ hereafter) of Cas12a complexes on DNA, we obtained the mean fluorescence signal over a 120 s time window (Figure 1B). To map the binding of Cas12a to base pair, the naturally occurring signal in the 488 channel (Supplementary Figure S2c) from the polystyrene 4.38 μm beads that indicate the bead's center was localized using a Gaussian filter. The center position was tracked across the whole 120 s window. The signal coming from the DNA tether region (Supplementary Figure S2c) was defined as the bead center-to-center distance minus twice the radius of the beads. The DNA’s 48.5 kb was then mapped to the DNA region. The error in fitting, introduced by system variability and the algorithm itself, was computed by determining the difference between the fluorescence intensity peak location and the designed on-target location after mapping. Using the 120 s-long profile, the center of the peak was found by using a gaussian fit. The error (or the difference between the on-target and the fluorescence peak) was found to be 232 ± 113 bp (n = 4; one profile was excluded from analysis due to the absence of on-target binding), where 113 is the standard deviation.
The background was defined as the minimal signal found in the region outside of the construct (Supplementary Figure S2c). Once the background was subtracted and the DNA distance was mapped in base pairs, a manual inspection of the binding profile was done to assess DNA orientation. The orientation of the DNA was determined based on alignment of the profile with the on-target site, recurrent off-target binding sites and with the typical asymmetrical binding preference that was consistently observed at higher forces. To facilitate data visualization and comparison between different kymographs, all profiles from a single protein were plotted on the same plot (grouped by force level). Each binding profile was further normalized to the highest intensity value present within any of the profiles. To pool all conditions into a single figure, an average was calculated over 500 bp windows at each force for each protein. These profiles were then plotted against the other forces and conditions in a single plot. To facilitate data visualization, each of these profiles were further normalized to the highest intensity value present within these profiles. For more information on how the DNA-basepair mapping is done with an example data set, see comparable analysis scripts available on Harbor (www.harbor.lumicks.com).
10s binding profile (binding dynamics)
To visualize binding dynamics, a single representative kymograph was selected. The 120sec kymograph was averaged in 12 × 10 s windows to allow detection of short timescale dynamic events. Profiles were grouped per force and plotted consecutively to show dynamic changes over time. Each profile was normalized to the highest intensity value present within profiles across the full force range.
Cleavage
Cleavage events were captured using dynamic or static assays. In both assays, the sudden decrease in force within the 200 s timeframe was manually identified through visual inspection. When no cleaving was observed before the 200 s timeframe, these cases were classified as non-cleaving. Force–extension curves showing the sudden decrease in force were obtained by plotting the force in the x-direction on Trap 2, downsampled to 15 Hz (from 78 kHz) against the camera-based distance tracking. The time-to-cleavage distribution for the WT, Cas12a_M1 and Cas12a_M2 on ssDNA was obtained using an automated change-point analysis detecting the sudden drop in force.
Off-target sequence mapping and data analysis
Cas12a has been shown to efficiently target spacer sequences following 5′T-rich PAM sequence. While the PAM for Lachnospiraceae bacterium (Lb) Cas12a and Acidaminococcus sp. (As) Cas12a has a sequence of 5′-TTTN-3′, FnCas12a has a 5′-TTN-3′ PAM. Therefore, it can also recognize 5′-TTTN-3′ (44). The theoretical off-target data analysis also included searches for short nucleotide sequences complementary to the crRNA (4–7 nts) starting at the PAM-proximal side of the crRNA selected and compared across the full λ-DNA sequence. A match on the λ-DNA is found if anywhere on the DNA, on either strand, the short sequences are identical and in the same order. Short sequences including the TTN PAM (7–10 bp) and the TTTN PAM (8–11 bp) were used as well, with the same number of bases post-PAM as the sequences without PAM but with an additional 3 (TTN PAM) or 4 (TTTN PAM) nts prior to the 4–7 PAM-proximal NT-strand. Searches were performed on both target strand and non-target strand, both in 5′→ 3′ direction. To determine variability in off-target location, a Gaussian fit was performed on the 120s-long profile at the observed main off-target binding event. For n = 4, the average peak location was found at 13.329 bp with a standard deviation of 197 bp. One profile was excluded from analysis due to an ambiguous main off-target binding event.
RESULTS
Cas12a binding and cleavage of λ-DNA
To monitor Cas12a–DNA interactions at the single molecule level we combined optical tweezers with confocal fluorescence microscopy and microfluidics (Figure 1a) (45,46). The unpredictable success of crRNA design and target selection is a still standing issue for the activity of CRISPR nucleases such as Cas9 and Cas12a, which usually results in designing 3–4 crRNAs to target a single gene (47,48). In vivo the target accessibility in a chromatin context could play an important role, but in vitro the secondary structure of the crRNA is also a major determinant of the crRNA cleavage performance (49,50), especially in the case of Cas12a, where the editing efficiency has been observed to depend on the design of the crRNA (51). To analyze Cas12a activity, we selected a location that is efficiently cleaved. Therefore, we screened crRNAs targeting five sites on λ-DNA and chose the sequence targeting the site where we observed a robust cleavage (Supplementary Figure S1, Table S1). Therefore, to proceed with the analysis, Cas12a was assembled with a 3′-Cy5-labeled version of crRNA-1 to form the ribonucleoprotein complex. Binding along the λ-DNA was visualized by recording kymographs from the Cy5 fluorescent label at the 3′ of the crRNA (Figure 1B, Materials and Methods). The selected crRNA-1 sequence binds a unique target site located at 29.7 kb on λ-DNA (Supplementary Table S1). The λ-DNA was attached to two polystyrene beads using the biotin-streptavidin affinity system and a force of 10 pN was applied to the λ-DNA pulling the beads. This force allows stretching of the λ-DNA without altering its base pairing (43,52). To use the wild-type Cas12a and avoid cleavage of the target DNA, we performed binding experiments with the crRNA-1/Cas12a complex in the absence of Mg2+. The assay showed Cas12a binding on dsDNA without cleaving the tether (Figure 1B). No binding was observed with 3′-Cy5-labeled crRNA-1 alone, confirming that the observed events represent binding of the assembled ribonucleoparticle (Supplementary Figure S2a). Next, we included Mg2+ in the buffer to observe Cas12a cleavage. The presence of the divalent metal ion triggered an immediate loss of tension on the tether, indicating the disruption of the DNA molecule (Figure 1C).
Off-target Cas12a binding correlates with tension
Cas12a binding kymographs were also obtained at forces from 10 to 50 pN (Figure 2A). The signal of the kymographs was averaged over a time-window to obtain fluorescent intensity profiles (Materials and Methods). The section of the kymograph corresponding to the DNA was identified using a bead detection algorithm and mapped to kb. The resulting on-target binding location on the mapped DNA was found to be within 232 ± 113 bp (mean and SD, n = 4) of the theoretical on-target site (Figure 2B, Supplementary Figure S2b, c, Materials and Methods). By using 10s-windows over the 120s-long kymograph, we obtained 12 intensity profiles per force, with each intensity profile showing the binding location and binding intensity averaged over a 10 s window (Figure 2C).
Figure 2.
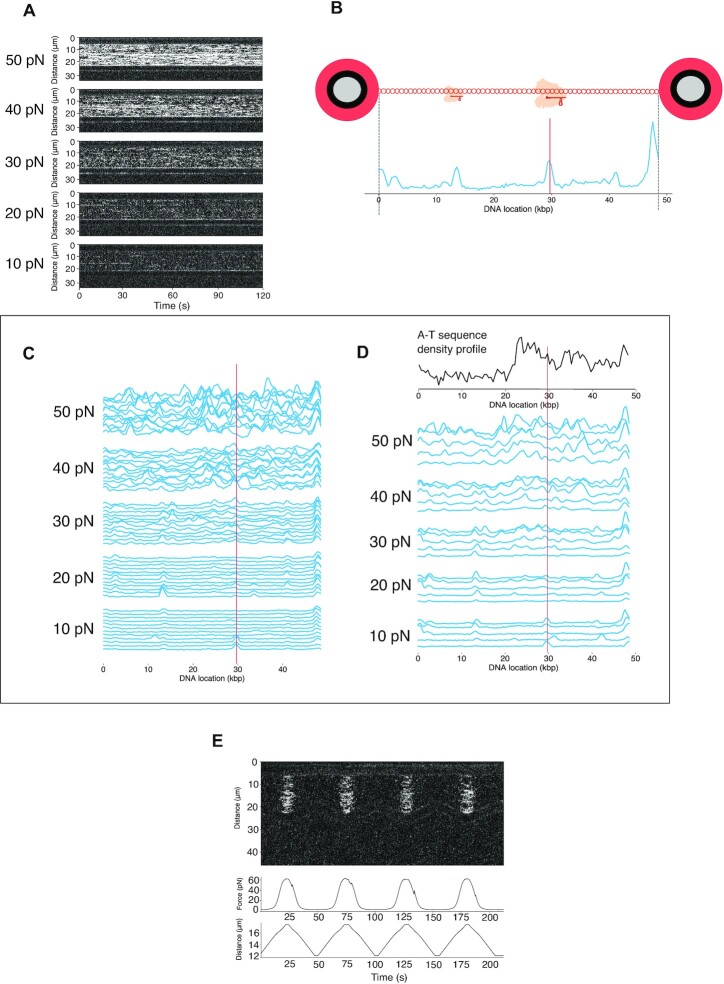
Kymographs and binding profiles of Cas12a with increasing tension. (A) Representative series of 120s kymographs, showing Cas12a binding with increasing forces (n = 5). (B) Schematic showing how profiles were generated. Line-scans were time-binned and cumulative fluorescence intensity was plotted along the DNA axis. (C) A single representative kymograph was binned 10s at each force, visualizing the dynamic nature of binding events. The red line indicates the target site centered at 29.707 kb, which is at the middle of the target sequence including a TTTN PAM. (D) Repeats of binding profiles at increasing forces binned at 120 s (n = 5). The red line indicates the target site centered at 29.707 kb. The A–T density profile along the λ-DNA is indicated at the top of the plot. (E) Representative kymograph and force trace of a repeated force-distance cycling (n = 15).
Cas12a displayed steady binding to λ-DNA over several 10 s intervals and shorter binding events when a 10 pN tension was applied on the DNA molecule (Figures 1B and 2A–C). Sometimes after initial binding at a given site, the crRNA-1/Cas12a complex becomes no longer visible, suggesting a wide range in binding durations over the entire tether. This could potentially be explained by the dissociation and rebinding of crRNA-1/Cas12a on a given site, as it cannot complete cleavage due to the absence of Mg2+. At high forces, between 30 and 50 pN, the increase of tension in the DNA molecule facilitates the binding of the ribonucleoprotein in multiple sites, indicating off-target binding events (Figure 2C). A few of these off-target binding events are more recurrent and persist across different forces. Alternatively, by using a single 120 s-long window, we obtained a single intensity profile per force (Figure 2D). The resulting intensity profiles displayed different binding sites with an overall higher tendency to locate in the AT-rich regions of the λ-DNA (Figure 2D), in contrast to Cas9 which shows a GC region preference (53). The correlation between increased tension and increased binding is highly reproducible. Indeed, reducing the force to 10 pN results in the dissociation of most complexes from the DNA (Figure 2E). This behaviour agrees with the PAM preference of Francisella novicida (Fn) Cas12a (TTN/TTTN) and the PAM scanning stage of the proposed model of PAM-dependent DNA recognition and ATP-independent unwinding at low forces (24). However, at high forces, when the melting of the tether generates ssDNA, binding of the RNP seems to circumvent the PAM recognition and is favoured by partial Watson-Crick pairing with the crRNA.
Cas12a off-target binding aligns with predicted PAM locations
Next, we investigated Cas12a potential off-target binding locations by analysing the λ-DNA sequence. We searched for partial matches with the crRNA that could potentially induce off-target binding with the crRNA, considering the PAM preference of FnCas12a (TTN/TTTN). We examined for Watson-Crick coupling using short nucleotide sequences complementary to the crRNA (4–7 nt) starting at the seed sequence of the crRNA-1 spacer across the full λ-DNA sequence (Materials & Methods, Supplementary Figure S3).
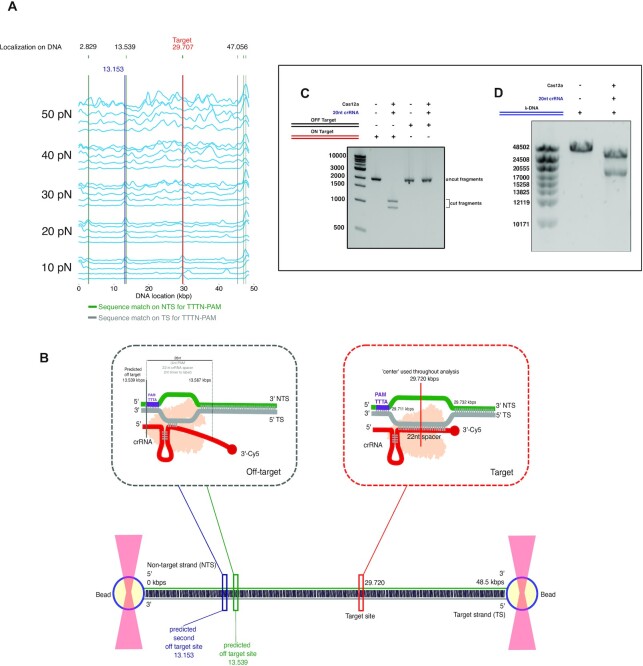
If we do not consider the PAM as a condition sine qua non for binding, the analysis shows that there is a total of 251 different sites on λ-DNA that match with at least 4-nt of the crRNA sequence, including the target site (Supplementary Figure S3a). However, PAM recognition is an indispensable preliminary event for Cas12a target DNA cleavage (10). Therefore, we performed a similar analysis considering the TTN and TTTN PAM sequences. Including TTN and 4-nts immediately after the PAM, the number of matching sites is reduced to 29 (Supplementary Figure S3b), and further reduced to 8 matching sites (including the on-target) when using TTTN (Figure 3A). Interestingly, the predictions of the off-target binding sites based on the PAM location and the adjacent sequence complementarity correlates strongly with the mapping of the experimental binding profile (Figure 3A, Supplementary Figure S3a, b), suggesting that the few consistent off-target binding events of Cas12a are not random but directed by Watson–Crick coupling, which is favoured at higher tensions. One main off-target site experimentally found at 13.329 ± 0.197 kb (mean and SD, n = 4) (see Materials & Methods) shows a particularly persistent binding across different forces. Interestingly, from our sequence analysis of the λ-DNA two off-target matches, 13.539 kb (matched using TTTN-PAM) and 13.153 kb (matched using TTN-PAM) lie in very close proximity to the main off-taret binding site at 13.329 ± 0.197 kb (determined by confocal bp localisation) (Figure 3A). Another persistent off-target binding site at ∼48 kb, although correlating very well with two nearby sequence-matched sites (Figure 3A), was too close to the bead edge for us to confidently separate the signal resulting from the bead proximity from the one resulting from the binding on the DNA. As such, that site was not investigated further.
Figure 3.
Analysis of predicted and observed off-target binding sites. (A) Binding profile of Cas12a (120 s binned, n = 5) showing sequence matches to TTTN-PAM on NT-strand (5′→ 3′, green lines) and on T-strand (5′→ 3′, grey lines). The blue line indicates a single sequence match to TTN-PAM on the NT-strand (Supplementary Figure S3a, b for all matches). (B) Scheme showing tentative positions of predicted and observed binding sites along the DNA. (C) 2% agarose gel showing Cas12a enzymatic activity on ∼2000-bp fragments of the λ-DNA containing the target and the off-target sites (n = 3). (D) 0.7% agarose gel showing the enzymatic activity of Cas12a on the complete λ-DNA (n = 3)
To evaluate whether this off-target binding event at 13.329 kb could lead to a DSB using crRNA-1, we performed a cleavage assay using first a target DNA of 2000 bp including the 13.153 kb and 13.539 kb off-target binding sites and tested the cleavage activity of the crRNA-1/Cas12a complex in the presence of Mg2+. In parallel, the experiment was performed with another 2000 bp DNA containing the target site at 29.720 kb (Figure 3C, Supplementary Table S1). Cleavage could not be detected in the off-target binding sites, while the crRNA-1/Cas12a complex efficiently generated a DSB in the selected target DNA. To further evaluate whether any of the other binding events could lead to unspecific cleavage, we performed a similar activity assay using the complete λ-DNA as substrate (Figure 3D). The linear molecule was incubated with the crRNA-1/Cas12a in the presence of Mg2+ under the previous conditions. Cleavage was only observed at the selected site targeted by crRNA-1, indicating that despite the observed off-target binding, the crRNA-1/Cas12a complex only generates a DSB at the selected site.
Collectively, these results suggest that Cas12a performs a PAM-guided scanning of the possible binding sites and tests complementarity with the guide (24). However, the target DNA is severed only when the PAM and the subsequent crRNA/T-strand hybrid requirements have been fulfilled (25).
DSB on λ-DNA is observed at low forces
Next, we analyzed the cleavage of Cas12a on λ-DNA. We performed an optical tweezers experiment stretching the DNA tether in the presence of Mg2+, thus allowing specific hydrolysis of the phosphodiester bonds (23,24,37). The tension applied to the λ-DNA was progressively increased from 0 to 65 pN during 50 s, after which the DNA molecule was relaxed to lower force conditions (Figure 4A). This cycle was repeated up to four times, and a tether breaking before the four cycles was considered a DSB initiated by the Cy5-crRNA-1/Cas12a complex. The break of the tether is detected by a sudden drop in force (Figure 4B), and no DSB events were observed without Mg2+ (Figure 1C). Detection of the DSB by Cas12a was exclusively observed ≤5 pN of tension, either on the extension or relaxation steps (Figure 4C), suggesting that higher tension is not required for DSB generation nor for the release of the cleaved DNA, as previously observed for Cas12a (54).
Altogether, our binding and cleavage experiments suggest that Cas12a explores possible target sites, and phosphodiester hydrolysis to generate the DSB, which in contrast with Cas9 whose cleavage is visualized at a substantial mechanical force (40 pN) (53), is observed at the selected target site at low forces. This observation suggests that Cas12a needs a flexible DNA to generate the break on the tether.
Cas12a variants binding to dsDNA
After dissecting wtCas12a mechanical properties during target DNA recognition and cleavage, we analysed and compared three engineered variants with different cutting properties. The fact that Cas12a displays an indiscriminate ssDNA degradation activity, which is observed in all its orthologs (28), together with recent studies that reported non-specific nicking of target sequences bearing mismatches in distal regions of the target DNA (55), suggest that these cutting activities could be a problem for potential applications. To overcome these problems, we have engineered three variants of Cas12a with singular cleavage properties (25).
These variants displayed different cleavage patterns in activity assays using the crRNA (Supplementary Table S1 crRNA-6), which was used in in our previous study (25) (Supplementary Figure S4a). The variants depicted similar cleavage patterns when the RNP was assembled with crRNA-1 (Figure 5A, B). This RNP was subsequently used to dissect their binding and cleavage properties in the optical tweezers. These variants are: (i) the REC-Sub mutant (Cas12a_M1) (25), which only displays unspecific ssDNA cleavage after activation with at least a 16–17 nt ssDNA oligonucleotide complementary to the guide, hereafter termed activator; (ii) the Q1025G/E1028G mutant (Cas12a_M2), whose ssDNA unspecific cleavage activity has been abolished and only cuts dsDNA specifically and (iii) the K1013G/R1014G mutant (Cas12a_M3), which presents a specific nickase activity, as it displays preferential phosphodiester cleavage on the T-strand of the target DNA. In addition, we included in the assays the E1006A-D917A catalytically dead mutant (Cas12a_KO) as a control (Figure 5A, B).
Figure 5.
Cas12a variants biochemical cleavage assay and binding on dsDNA and ssDNA. (A) 15% urea gel showing enzymatic activity of Cas12a with crRNA-1 (See Supplementary Table S1) with the annealed dsDNA formed by TS-1 and NTS-1 (see Supplementary Table S1 and Materials and Methods). (n = 3). (B) 15% urea gel showing indiscriminate ssDNA activity of Cas12a variants using crRNA-1, and activated by Activator TS (See Supplementary Table S1), with Unspecific ssDNA (See Supplementary Table S1) as the substrate (n = 3). (C) Binding profiles of wtCas12a and its variants at increasing forces binned at 120 s (n = 5, at each force for all the samples). The red line indicates the target site centered at 29.720 kb, which is located at the middle of the target sequence including a TTTN PAM. The black line indicates the prominent off-target binding site at 13.539 kb. (D) Representative 60 s-long kymographs showing binding of wtCas12a and its variants to ssDNA in the presence and absence of the activator at a force of 20 pN. All kymographs were obtained in the presence of Mg2+. In addition, in experiment the ssDNA tether was first exposed to a buffer without the activator, then, the same tether was moved to the buffer with the activator. In the case of wtCas12a and Cas12a-M1, the kymographs show a section of the data prior to the breaking of the tether.
The experiments were performed in the same conditions as in Figure 2A (Materials and Methods). Overall, all the mutants show a wide range in binding durations over the entire λ-DNA like wtCas12a, displaying only small variations between repetitive kymographs. Overall, the binding profiles also depict an on-target site at 29.707 kb and the main off-target site around 13.000 kb (Figure 5C, Supplementary Figure S4b, c). However, some variations can be observed at 10 pN, reflecting differences in binding. However, when tension is applied to the tether the two peaks are shown but with different intensities. For instance, in the case of Cas12a_M2 the intensity of the binding sites is small at 10 pN and it is observed more clearly at 30 pN. This is similar to Cas12a_M1, which displays a peak around 13.000 kb at lower forces and displays the second binding location at 29.707 kb when tension between 20 and 30 pN is applied to the tether. A similar behaviour can be observed for Cas12a_KO, which binds only to on-target at 29.707 kb at low forces and shows the binding event around 13.000 kb when force is applied to the DNA. At high forces (40–50 pN) unspecific binding starts to predominate. Besides the main sites, all of them also display a preference to bind the AT-rich region of the λ-DNA at high forces (Supplementary Figure S4b, c). Collectively, we observed that all the variants show a dsDNA binding pattern similar to the wild type. However, differences could be observed in the peak intensity, suggesting that while in general their behaviour is conserved, there could be differences in target affinity.
Cas12a variants binding to ssDNA
As unspecific cleavage can be unleashed after specific DSB generation (28), we investigated in detail the binding of M1 and M2 on ssDNA, as we dissociated in these variants the specific dsDNA cleavage and the unspecific ssDNA degradation after activation (Figure 5A, B) (25). For that purpose, we tested binding to ssDNA using the λ-DNA strand corresponding to the ‘T-strand’ as tether in the presence and absence of an activator, a complementary ssDNA of 40-nt, at 20 pN (Figure 5D, Supplementary Table S1). For wtCas12a, we did not observe binding on ssDNA in the absence of the activator. However, when the activator is present multiple binding events on the ssDNA can be observed in the 60 s kymograph, suggesting that assembly of the hybrid between the crRNA and the activator facilitates the interaction with the ssDNA by opening the lid in the RuvC domain (25). A similar behaviour can be observed for Cas12a_M1, which shows a slightly higher level of binding without the activator compared to the wild type but displayed a remarkably high number of binding events when the activator was present. This indicates that the mutations in Cas12a_M1 promote ssDNA binding in the presence of activator. On the other hand, the Cas12a_M2 mutant shows no binding to ssDNA without activator and a minimal binding when the activator is present. The observed Cas12a_M1 and Cas12a_M2 binding behaviour supports the bulk activity assays (Figure 5A, B), showing that Cas12a_M1 preferentially cuts ssDNA while Cas12a_M2 is unable to unleash the unspecific ssDNA activity, as it also does not bind ssDNA.
Collectively, the ssDNA binding behaviour of these variants supports the idea that the hybrid assembly between the crRNA-1 and the activator promote the conformational change that opens the ‘lid’ in the catalytic pocket (25), allowing binding to the ssDNA to hydrolyse the phosphodiester bonds. By contrast binding on ssDNA is not observed for the Cas12a_M2, whose unspecific ssDNA cleavage activity has been eliminated by the double mutation and only cleaves dsDNA targets.
Cas12a variants cleavage on ds- and ssDNA
To analyze the cleavage activity on ds- and ssDNA, we subjected the variants to two types of experiments: A dynamic assay in which the dsDNA tether was repeatedly stretched from 0 to 65pN for up to four times (Figure 6Aa), and a static format in which the ds- or ssDNA tether was maintained at 20 pN for 200 s (Figure 6B). The assays were performed in the presence or absence of activator depending on the experiment and Mg2+ was included in these experiments (Figure 5A, B). The static cleavage assays for variants Cas12a_KO and Cas12a_M3 were not performed, as Cas12a_KO does not display catalytic activity and Cas12a_M3 activity of can only be observed at high forces. In addition, neither of these variants display the indiscriminate ssDNase activity.
Figure. 6.
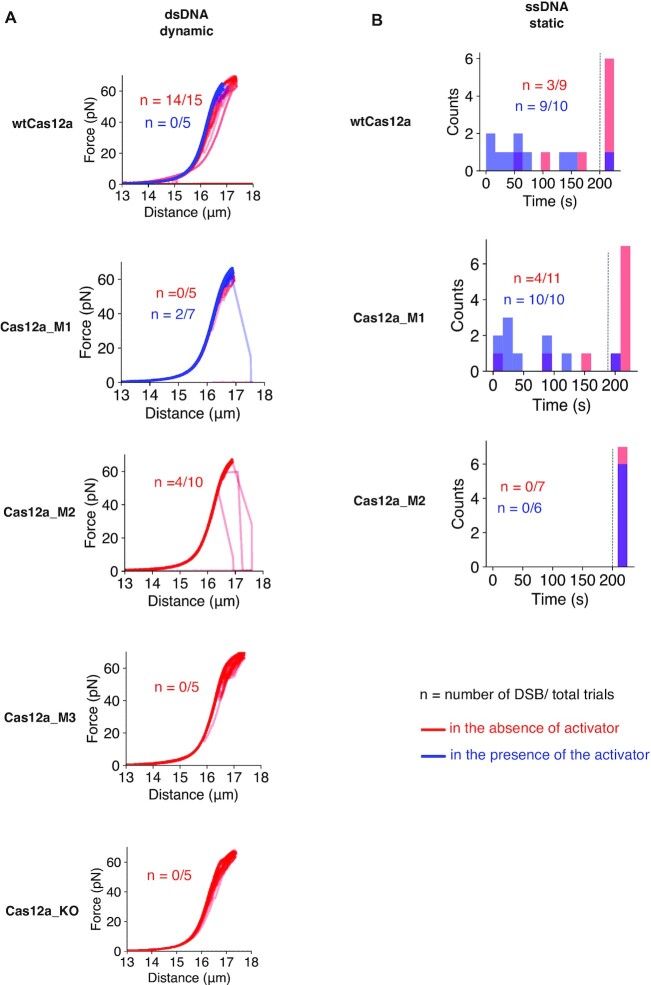
Quantitative analysis of dsDNA and ssDNA cleavage by wtCas12a and its variants. (A) Force-extensions curves of λ-DNA tethers in the presence and Mg2+ and wtCas12a and its variants for dynamic dsDNA cleavage experiments. (B) Plots of unspecific ssDNA degradation in a static setting by wtCas12a and its variants as a distribution of time. All experiments in the static form were conducted at 20 pN. λ-DNA ssDNA tethers that remained present after 4 force–distance cycles (A, dynamic) or 200s (B, static) were considered non-cleaved (indicated by the dashed line). The static cleavage assays for variants Cas12a_KO and Cas12a_M3 were not performed, as Cas12a_KO does not display catalytic activity and Cas12a_M3 activity of can only be observed at high forces. In addition, neither of these variants display the indiscriminate ssDNase activity.
The generation of a DSB in the tether by wtCas12a is hampered by the presence of the activator. However, the activator enhances and speeds up the cut on the ssDNA tether (Figure 6A, B), suggesting that the formation of the hybrid between the crRNA and the activator disturbs the recognition of the target site on the double-stranded λ-DNA and thereby cleavage. Yet the formation of the hybrid with the activator opens the catalytic RuvC/Nuc pocket, promoting the unspecific binding (Figure 5D) and cutting of the ssDNA (25).
Single molecule experiments also supported the observation that the Cas12a_M3 variant is unable to cleave dsDNA. No loss of tension was observed in the dsDNA tether when this mutant was tested (Figure 6A). However, the use of Sytox orange, a DNA intercalator binding only to dsDNA (56,57), supported its nickase activity, since the absence of this specific dye is indictive of ssDNA, suggesting a possible peeling of the DNA upon tension, and thereby pointing towards severing of one of the strands, generating a region of ssDNA (Supplementary Figure S4d).
The binding analysis in the variants Cas12a_M2 and Cas12a_M1, where the unspecific ssDNA and specific dsDNA severing activities have been dissociated, show substantial differences. The bulk activities observed for these variants are well explained by our single molecule experiments. The dsDNA cleavage of Cas12a_M1 in the presence or absence of the activator is severely affected in the dynamic experimental setup. However, some cleavage can be observed at high forces, coinciding with the tension where ssDNA is available (Figure 6A). Therefore, the detected DSBs could be due to nicks on both strands of the λ-DNA, as in these experiment Cas12a_M1 is engaged with the activator. Like the wild type, Cas12a_M1 can partially cleave the ssDNA tether in absence of the activator (Figure 6B). The presence of the activator enhances this activity in wtCas12a while it triggers full activity faster for Cas12a_M1. All ssDNA molecules are severed within a wide distribution of time (Figure 6B). On the other hand, the ssDNA cleavage activity is completely abolished in the Cas12a_M2, even when the activator is present. This variant solely cuts dsDNA, but its cleavage activity is reduced (Figures 5A, B and 6A, B), suggesting that abrogation of the unspecific ssDNA activity affects the catalytic site.
In contrast with wtCas12a, the DSBs produced by Cas12a_M2 on the λ-DNA were observed at high forces around 50 pN (Figure 6A). The activity assay of Cas12a_M2 suggested that the T-strand is preferentially cut instead of the NT-strand (Figure 5A). Therefore, to test this hypothesis, we performed an assay to determine whether the substitutions, besides eliminating unspecific ssDNA activity, reverted the cleavage preference of this mutant (Supplementary Figure S5). The assay showed that the Cas12a_M2 variant cuts the T-strand faster, suggesting that the redesign favours a conformation that disturbs phosphodiester hydrolysis in the NT-strand. This change in cleavage preference suggests that the tension in the DNA is necessary to permit the conformational change, allowing NT-strand cleavage, and thereby visualisation of the dsDNA disruption. The preference to cleave the T-strand and not the NT-strand also suggests why this variant does not support unspecific ssDNA activity, we speculate that the entrance of the oligonucleotide would not be favoured by the conformation induced by the mutations. Therefore, our single molecule experiments provide a mechanical background to explain its different ss- and dsDNA severing activities.
DISCUSSION
Structure–function studies of Cas12a provided important snapshots of the molecular details of Cas12a (24–26,37,38), yet a mechanical characterization of the target recognition and cleavage by Cas12a and a comparison with some of its engineered variants to fully understand this novel endonuclease was lacking.
Cas12a versus Cas9
Our study reveals that Cas12a and the variants bind and dissociate from the λ-DNA while searching for the target sites at different locations where a complete or cryptic sequence (including partial matches with the crRNA) is found (Figures 2A and 7A). A previous study using AsCas12a has suggested that the RNP may slide several kb on random DNA binding when searching for the target sequence. Sliding was observed in 50% of the traces and is favored at higher ionic strength conditions in the presence of 100 mM Na+ and 10 mM Ca2+, till the RNP associates to the target sequence (58). We have not observed this behavior with FnCas12a in our experimental conditions, which include 150 mM KCl with or without 5 mM Mg2+. Besides the buffer, other differences between both studies are the location of the target site and the fluorescent probe in the RNA. We attached the probe in the 3′of crRNA-1 for FnCas12a, while in the study for AsCas12a the crRNA was designed with a modified T inside the handle of the crRNA, which is later coupled with the probe (58). Nevertheless, besides this difference on the initial binding, the analysis of As and FnCas12a are in good agreement in the rest of the events occurring after recognition of the target sequence leading to cleavage at low forces.
Figure 7.
Schematic model of Cas12a target binding and cleavage on λ-DNA. (A) The Rec 1and 2 subdomains are depicted in blue and orange while the NUC lobe is in grey and the crRNA in red. The active site is depicted with an oval which is colored in black with a yellow circle when is activated. Cas12a searches for its target DNA in a PAM dependent fashion. A lack of complementarity with the guide of the crRNA induces dissociation and the initiation of a new search for the target DNA. Once the target is found and the RNA/DNA hybrid is assembled catalysis proceeds in the NT-strand and later in the T-strand after a conformational change that positions it the catalytic site, thus leading to a specific DSB at low forces. At high forces the DSB is not observed as the high tension of the DNA hinders the conformational change that positions the T-strand in the active site. (B) The use of a ssDNA tether shows that only in the presence of an activator the assembly of the hybrid opens the ‘lid’ of the active site to unleash indiscriminate ssDNA cleavage. As the ssDNA is not under tension the possible cryptic binding by complementarity with crRNA is not favoured. The system may bind transiently to possible PAMs but it will not find the complementary strand to promote the phosphate inversion and test binding with the crRNA spacer.
We consistently observe a preference to bind onto AT rich regions where the PAM and PAM-like sequences are more frequent, confirming that PAM recognition is driving the search for the target DNA. The number of off-target sites observed for Cas12a increases when tension is applied to the λ-DNA (Figures 2A–C and 7B), as the crRNA may associate with exposed ssDNA cryptic sites. Two main binding locations are observed. The on-target site and one main off-target at 13.329 (excluding a secondary off-target site towards the end of the DNA and close to a polystyrene bead) (Figure 2A). Overall, the lifetime of the binding events is short, but these events are longer and more persistent in the target site and the main off-target site sharing the TTTN/TTN PAM and 4-nt of the guide with the target site (Figure 3A). After identifying the two preferential off-target binding sites based on sequence matching, the cleavage experiments using crRNA-1 in that region showed that binding of Cas12a did not lead to cleavage of dsDNA containing these two cryptic off-target sites. Furthermore, when a similar cleavage assay was performed with the complete λ-DNA, a single cut corresponding to the selected target site could be observed (Figure 3C and D). The optical tweezers experiments on λ-DNA revealed that Cas12a displayed a DSB at low forces ≤5 pN (Figures 4A, C and 7A), suggesting that it is unable to hold the ruptured DNA together. This agrees with previous observation indicating that the PAM distal site dissociates from the complex, while the PAM proximal site remains bound forming an R-loop (24,37,54). In addition, our binding and cleavage experiments with wtCas12a using a ssDNA tether in the presence of an activator support the conformational change caused by the crRNA/DNA assembly to open the catalytic site and promote its unspecific ssDNA binding and cleavage activity (Figures 6B and 7B).
Although Cas9 and Cas12a share some degree of structural organization (59), their binding and cleavage behavior on λ-DNA are different (53). Cas9 contains two active sites, one in the RuvC and other in the HNH domain and cleavage occurs next to the PAM. In contrast, the PAM binding site of Cas12a is located 70 Å away from the single RuvC/Nuc pocket where catalysis occurs. This structural organization suggests that in Cas9, PAM recognition and catalysis of the blunt end DSB are closely associated, while in Cas12a PAM recognition is physically separated from the single RuvC/Nuc active site to generate the overhang. Hence, the event of PAM scanning and binding is separated from the active site and needs the completion of at least a 16–17 nt long crRNA/T-strand hybrid to activate catalysis (25). Furthermore, RuvC domains preferentially cut DNA with the 5′-3′polarity (60), suggesting that a conformational change should occur after the cleavage of the NT-strand to position the T-strand in the RuvC catalytic pocket for cleavage (25). These structural differences suggest that Cas9 would generate DSBs on the λ-DNA when tension is applied to the tether (53), as ssDNA is available and the HNH and RuvC domains can catalyze the phosphodiester hydrolysis in both strands.
However, in the case of Cas12a, the break on the λ-DNA is visualized at low forces ≤5 pN. This observation suggests that the T-strand would need some plasticity to enter the RuvC catalytic site with the 5′→3′ polarity (Figures 4A, C and 7A). In addition, it would also support the notion that cleavage of the T-strand would release the tension in the target DNA, and the two halves would snap back and become flexible, opening the cavity between the REC and NUC lobes, unzipping the crRNA-DNA hybrid and allowing the entrance of the T-strand in the catalytic pocket for catalysis (25). The lack of DSB detection at high forces suggest that the proposed conformational change would be hindered at high forces, as under high-tension the stretched T-strand would not be inserted with a 5′-3′polarity in the RuvC/Nuc pocket (Figure 7B). However, in the case of Cas9, each strand is severed by a different catalytic center (1).
Collectively, these observations suggest that the numerous off-target binding events of Cas9 at high forces combined with its two catalytic centers and the stretched DNA conformation would favor the off-target cleavage on λ-DNA (53). By contrast, Cas12a can bind numerous cryptic-sites (Figure 7B), and it may slide (58), but the conformational restrictions due to its single catalytic pocket would prevent these off-target binding events at medium and high forces (Figure 2A–C) leading to breaks on the DNA (Figure 4A–C).
Cas12a variants
The analysis of the three Cas12a variants shows that they all conserve the binding behaviour to dsDNA similar to the wild type (Figure 5C, Supplementary Figure S4a, b, Figure 8). Their off-target binding increased when force was applied to the λ-DNA, and all of them show a more persistent association to the target, displaying a similar preference as the wild type for the off-target binding sites. This short-lived binding and the longer lifetime on the target and cryptic target sites indicate that PAM scanning and initial unzipping to test hybridisation are not affected by the changes introduced in these mutants. However, the variants display a different cleaving capacity in the bulk experiments, which is reflected in the single molecule experiments (Figures 5, 6 and 8).
Figure 8.
Schematic model of Cas12a_M1, M2 and M3 variants binding and cleavage on λ-DNA. Cas12a_M1 contains mutations in the linker between Rec1 and 2 and both are colored in green. In a ssDNA or dsDNA in the absence of activator, there is no cleavage either at low or high forces. Once the activator is present cleavage is observed in the case of the ssDNA tether and as well in the dsDNA in a minor extent. The mutations in the active site of Cas12a_M2 are depicted by a yellow oval. By contrast with Cas12a_M1, the Cas12a_M2 variant does not present unspecific ssDNA cleavage in the favorable conditions of Cas12a_M1 in the presence of activator. Only specific dsDNA cleavage is observed, but at high tension, in contrast with wtCas12, as the preference of cleavage is reversed in this variant (Supplementary Figure S5). In Cas12a_M3, the mutations are depicted by a yellow oval with a red border. The mutant does not cleave or bind ssDNA and it cleaves the T-strand of the dsDNA tether (Figure 5a-b, Supplementary Figure S4c).
Both the wild type and Cas12a_M2 cleave dsDNA, while Cas12a_M1 shows a minor DSB generation at high forces that may be due to its action on the ssDNA generated by the tension (Figure 8). We have eliminated the indiscriminate ssDNA cleavage in the Cas12a_M2 variant by inverting wtCas12a cleavage preference (Supplementary Figure S5). Cas12a_M2 does not cut ssDNA non-specifically (Figures 6 and 8). Therefore, this variant could potentially eliminate possible sources of off-targets in genome editing applications. However, the deletion of this collateral activity of Cas12a seems to affect its catalytic center as the mutant displays a reduced cleavage efficiency of dsDNA. Consequently, an optimization of its catalysis maintaining its cleavage properties is needed to improve its efficiency.
The Cas12a_M3 binds to dsDNA but it does not generate a DSB on the target and it does not display indiscriminate ssDNA activity (Figure 5A, B). However, this variant is a sluggish enzyme as cleavage is not observed throughout the 200 s of the dynamic experimental setup. The activator-mediated cleavage of ssDNA in wtCas12a and Cas12a_M1 correlates well with the observed binding and cleaving activity. Binding to ssDNA and cleavage is enhanced, especially in Cas12a_M1, in the presence of the activator in agreement with the activation model of catalysis after hybrid assembly (25). In addition, cleavage shows a wide distribution in time, supporting the unspecific character of this activity.
The high conservation of these regions and the mutated residues (25), suggests that similar redesigns in other Cas12a orthologues should render similar cleavage activities; however, this needs to be tested experimentally. These redesigned Cas12a variants could have applications in different biomedical aspects. In that regard, the Cas12a_M1 variant is an improved version of the unspecific ssDNA activity displayed by wtCas12a which has been shown to be useful in diagnostics (28). For therapeutic genome editing applications, we foresee the Cas12a_M2 and M3 variants as promising candidates. In the case of Cas12a_M2, the indiscriminate ssDNA activity is eliminated, suggesting that this variant could display less toxicity than the wild type during editing, as it should not cleave ssDNA during editing. For Cas12a_M3, which is to our knowledge the first Cas12a nickase, base editing applications could be envisioned, as nicking in the selected region could lead to base conversion (61).
Our work shows that the combination of mechanical information using optical tweezers with structural and biochemical data can render valuable information to understand CRISPR-Cas endonucleases that may be later used in their redesign for in cellular and in vivo genome editing approaches.
DATA AVAILABILITY
All data are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
Guillermo Montoya is member of the Integrative Structural Biology Cluster (ISBUC) at the University of Copenhagen. We thank Janna Bogers (LUMICKS) for her helpful assistance in C-Trap experiments. We thank N. Hatzakis for careful reading of our manuscript.
Contributor Information
Bijoya Paul, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, Copenhagen 2200, Denmark.
Loïc Chaubet, LUMICKS, Pilotenstraat 41, 1059 CH, Amsterdam, The Netherlands.
Dideke Emma Verver, LUMICKS, Pilotenstraat 41, 1059 CH, Amsterdam, The Netherlands.
Guillermo Montoya, Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, Copenhagen 2200, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation [NNF14CC0001]; Distinguished Investigator [NNF18OC0055061 to G.M.]. Funding for open access charge: NNF [14CC0001, NNF18OC0055061].
Conflict of interest statement. Guillermo Montoya declares that is a co-founder of Twelve Bio and coauthor in a patent filing including the variants.
REFERENCES
- 1. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-rna–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cong L.e.a. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mojica F.J.M., Diez-Villasenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 5. Horvath P., Barrangou R.. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010; 327:167. [DOI] [PubMed] [Google Scholar]
- 6. Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A.. Cas13d is a compact RNA-Targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018; 70:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. eLife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K.. Genome-wide specificities of CRISPR-Cas cpf1 nucleases in human cells. Nat. Biotechnol. 2016; 34:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim D., Kim J., Hur J.K., Been K.W., Yoon S.-h., Kim J.-S. Genome-wide analysis reveals specificities of cpf1 endonucleases in human cells. Nat. Biotechnol. 2016; 34:863–868. [DOI] [PubMed] [Google Scholar]
- 10. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A.et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ernst M.P.T., Broeders M., Herrero-Hernandez P., Oussoren E., van der Ploeg A.T., Pijnappel W.W.M.P.. Ready for repair? Gene editing enters the clinic for the treatment of human disease. Mol. Ther. Methods Clin. Dev. 2020; 18:532–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V.et al.. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019; 10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M., Bassel-Duby R., Olson E.N.. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017; 3:e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee J.G., Ha C.H., Yoon B., Cheong S.-A., Kim G., Lee D.J., Woo D.-C., Kim Y.-H., Nam S.-Y., Lee S.-w.et al.. Knockout rat models mimicking human atherosclerosis created by Cpf1-mediated gene targeting. Sci. Rep. 2019; 9:2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ming M., Ren Q., Pan C., He Y., Zhang Y., Liu S., Zhong Z., Wang J., Malzahn A.A., Wu J.et al.. CRISPR–Cas12b enables efficient plant genome engineering. Nat. Plants. 2020; 6:202–208. [DOI] [PubMed] [Google Scholar]
- 18. Ren Q., Sretenovic S., Liu S., Tang X., Huang L., He Y., Liu L., Guo Y., Zhong Z., Liu G.et al.. PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat. Plants. 2021; 7:25–33. [DOI] [PubMed] [Google Scholar]
- 19. Pausch P., Al-Shayeb B., Bisom-Rapp E., Tsuchida C.A., Li Z., Cress B.F., Knott G.J., Jacobsen S.E., Banfield J.F., Doudna J.A.. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020; 369:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z., Zhang H., Xiao R., Han R., Chang L.. Cryo-EM structure of the RNA-guided ribonuclease cas12g. Nat. Chem. Biol. 2021; 17:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A.. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018; 362:839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karvelis T., Bigelyte G., Young J.K., Hou Z., Zedaveinyte R., Budre K., Paulraj S., Djukanovic V., Gasior S., Silanskas A.et al.. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020; 48:5016–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V.et al.. Crystal structure of cpf1 in complex with guide RNA and target DNA. Cell. 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stella S., Alcon P., Montoya G.. Structure of the cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017; 546:559–563. [DOI] [PubMed] [Google Scholar]
- 25. Stella S., Mesa P., Thomsen J., Paul B., Alcon P., Jensen S.B., Saligram B., Moses M.E., Hatzakis N.S., Montoya G.. Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell. 2018; 175:1856–1871. [DOI] [PubMed] [Google Scholar]
- 26. Swarts D.C., Jinek M.. Mechanistic insights into the cis- and trans-acting DNase activities of cas12a. Mol. Cell. 2019; 73:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nalefski E.A., Patel N., Leung P.J.Y., Islam Z., Kooistra R.M., Parikh I., Marion E., Knott G.J., Doudna J.A., Le Ny A.M.et al.. Kinetic analysis of cas12a and cas13a RNA-Guided nucleases for development of improved CRISPR-Based diagnostics. iScience. 2021; 24:102996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A.. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018; 360:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R.. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell. 2018; 71:816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S.et al.. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 2017; 35:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L., Wei K., Zheng G., Liu X., Chen S., Jiang W., Lu Y.. CRISPR-Cpf1-Assisted multiplex genome editing and transcriptional repression in streptomyces. Appl. Environ. Microbiol. 2018; 84:e00827-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J.. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discovery. 2017; 3:17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S.K., Kim H., Ahn W.-C., Park K.-H., Woo E.-J., Lee D.-H., Lee S.-G.. Efficient transcriptional gene repression by type V-A CRISPR-Cpf1 from eubacterium eligens. ACS Synth. Biol. 2017; 6:1273–1282. [DOI] [PubMed] [Google Scholar]
- 34. Kim H., Kim S.-T., Ryu J., Kang B.-C., Kim J.-S., Kim S.-G.. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017; 8:14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fonfara I., Richter H., Bratovic M., Le Rhun A., Charpentier E.. The CRISPR-associated DNA-cleaving enzyme cpf1 also processes precursor CRISPR RNA. Nature. 2016; 532:517–521. [DOI] [PubMed] [Google Scholar]
- 36. Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J.. Multiplexed genome engineering by cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods. 2019; 16:887–893. [DOI] [PubMed] [Google Scholar]
- 37. Swarts D.C., van der Oost J., Jinek M.. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell. 2017; 66:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O.. Structural basis for the canonical and Non-canonical PAM recognition by CRISPR-Cpf1. Mol. Cell. 2017; 67:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pelz B., Žoldák G., Zeller F., Zacharias M., Rief M.. Subnanometre enzyme mechanics probed by single-molecule force spectroscopy. Nat. Commun. 2016; 7:10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat. Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Candelli A., Holthausen J.T., Depken M., Brouwer I., Franker M.A.M., Marchetti M., Heller I., Bernard S., Garcin E.B., Modesti M.et al.. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berg-Sørensen K., Flyvbjerg H.. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 2004; 75:594–612. [Google Scholar]
- 43. Smith S.B., Cui Y., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 44. Paul B., Montoya G.. CRISPR-Cas12a: functional overview and applications. Biomed J. 2020; 43:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gross P., Farge G., Peterman E.J.G., Wuite G.J.L.. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010; 475:427–453. [DOI] [PubMed] [Google Scholar]
- 46. Heller I. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat. Methods. 2013; 10:910–916. [DOI] [PubMed] [Google Scholar]
- 47. Chari R., Mali P., Moosburner M., Church G.M.. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015; 12:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uusi-Mäkelä M.I.E., Barker H.R., Bäuerlein C.A., Häkkinen T., Nykter M., Rämet M.. Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS One. 2018; 13:e0196238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chu V.T., Graf R., Wirtz T., Weber T., Favret J., Li X., Petsch K., Tran N.T., Sieweke M.H., Berek C.et al.. Efficient CRISPR-mediated mutagenesis in primary immune cells using crisprgold and a C57BL/6 cas9 transgenic mouse line. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12514–12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thyme S.B., Akhmetova L., Montague T.G., Valen E., Schier A.F.. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun. 2016; 7:11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W.et al.. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017; 14:153–159. [DOI] [PubMed] [Google Scholar]
- 52. Zhang X., Chen H., Le S., Rouzina I., Doyle P.S., Yan J.. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Newton M.D., Taylor B.J., Driessen R.P.C., Roos L., Cvetesic N., Allyjaun S., Lenhard B., Cuomo M.E., Rueda D.S.. DNA stretching induces cas9 off-target activity. Nat. Struct. Mol. Biol. 2019; 26:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alcon P., Montoya G., Stella S.. Assembly of francisella novicida cpf1 endonuclease in complex with guide RNA and target DNA. Acta Crystallogr. F Struct. Biol. Commun. 2017; 73:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murugan K., Seetharam A.S., Severin A.J., Sashital D.G.. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020; 295:5538–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Biebricher A.S., Heller I., Roijmans R.F.H., Hoekstra T.P., Peterman E.J.G., Wuite G.J.L.. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015; 6:7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yan X., Habbersett R.C., Yoshida T.M., Nolan J.P., Jett J.H., Marrone B.L.. Probing the kinetics of SYTOX orange stain binding to double-stranded DNA with implications for DNA analysis. Anal. Chem. 2005; 77:3554–3562. [DOI] [PubMed] [Google Scholar]
- 58. Losito M., Smith Q.M., Newton M.D., Cuomo M.E., Rueda D.S.. Cas12a target search and cleavage on force-stretched DNA. Phys. Chem. Chem. Phys. 2021; 23:26640–26644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stella S., Alcon P., Montoya G.. Class 2 CRISPR-Cas RNA-guided endonucleases: Swiss army knives of genome editing. Nat. Struct. Mol. Biol. 2017; 24:882–892. [DOI] [PubMed] [Google Scholar]
- 60. Wyatt H.D., West S.C.. Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 2014; 6:a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith C.J., Castanon O., Said K., Volf V., Khoshakhlagh P., Hornick A., Ferreira R., Wu C.T., Güell M., Garg S.et al.. Enabling large-scale genome editing at repetitive elements by reducing DNA nicking. Nucleic Acids Res. 2020; 48:5183–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.