Abstract
Establishing saturated mutagenesis in a specific gene through gene editing is an efficient approach for identifying the relationships between mutations and the corresponding phenotypes. CRISPR/Cas9-based sgRNA library screening often creates indel mutations with multiple nucleotides. Single base editors and dual deaminase-mediated base editors can achieve only one and two types of base substitutions, respectively. A new glycosylase base editor (CGBE) system, in which the uracil glycosylase inhibitor (UGI) is replaced with uracil-DNA glycosylase (UNG), was recently reported to efficiently induce multiple base conversions, including C-to-G, C-to-T and C-to-A. In this study, we fused a CGBE with ABE to develop a new type of dual deaminase-mediated base editing system, the AGBE system, that can simultaneously introduce 4 types of base conversions (C-to-G, C-to-T, C-to-A and A-to-G) as well as indels with a single sgRNA in mammalian cells. AGBEs can be used to establish saturated mutant populations for verification of the functions and consequences of multiple gene mutation patterns, including single-nucleotide variants (SNVs) and indels, through high-throughput screening.
INTRODUCTION
A large number of mutations involving insertions or deletions of multiple nucleotides (indels), as well as single-nucleotide variants (SNVs) associated with human genetic diseases (1–3) or traits of crops (4) and livestock (5) have been detected through high-throughput genome sequencing. However, the functions or consequences of most mutations, particularly SNVs, have not yet been determined. The establishment of saturated mutagenesis (6,7) in a specific gene through gene editing is an efficient approach for identifying the relationships between these mutations and the corresponding phenotypes.
CRISPR/Cas9-based sgRNA library screening (8–10), the method most frequently used to generate saturated mutant populations, often creates mutations consisting of insertions or deletions of multiple nucleotides and is therefore not applicable for identifying the functions of SNVs. The recently developed base editor (BE) system enables base substitution in the genome and is thus considered an effective tool for the generation of SNV-associated saturated mutant populations (11–13). The two early types of BEs, cytosine base editor (CBE) (14,15) and adenine base editor (ABE) (16), can perform only one type of base transition, i.e. C-to-T or A-to-G, respectively, which limits their utility in site-directed saturated mutagenesis. To address this limitation, a variety of new BEs, such as STEMEs (17), SPACE (18), Target-ACEmax (19), A&C-BEmax (20) and ACBE (21), have been created by fusing cytidine deaminase and adenine deaminase together with Cas9 nickase to concurrently achieve C-to-T and A-to-G conversions at the same target site in plant and mammalian systems. Nevertheless, the types of combinational variants introduced by these dual-editors do not meet the requirements of saturated mutagenesis. More recently, a new glycosylase base editor (CGBE), in which the uracil (U) glycosylase inhibitor (UGI) is replaced with U-DNA glycosylase (UNG), has been reported to efficiently induce not only targeted C-to-G base transversion but also C-to-T and C-to-A conversions (22–25). In addition, UNG in the CGBE system can excise the U-base created by the deaminase to form an apurinic/apyrimidinic (AP) site that initiates the DNA repair process, which introduces indel mutations via an error-prone repair mechanism (26). The C-to-T and C-to-A conversions as well as the indels generated by a CGBE are considered two unwanted by-products for precise base editing (22,23). Conversely, these by-products are believed to be an advantage when a CGBE is used to create a saturated mutagenesis population in a gene since they increase the diversity of base editing outcomes. Therefore, in this study, we fused a CGBE with another base editor that is able to induce A-to-G conversion, ABE, to produce a new type of dual deaminase-mediated base editor named AGBE. The AGBE simultaneously generated A-to-G and C-to-G/C-to-T/C-to-A conversions as well as indels and thus substantially extended the diversity of variants in the same DNA strand at the target sites (Figure 1).
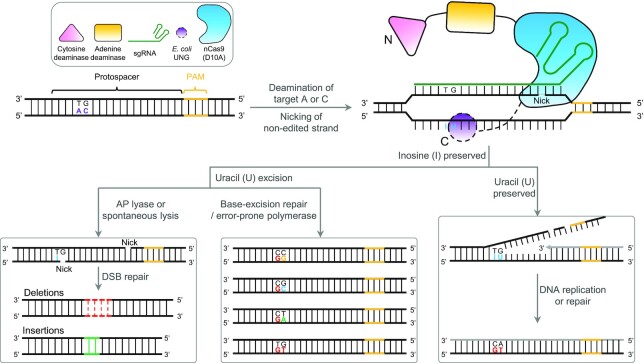
Figure 1.
Schematic of potential cellular mechanisms and outcomes of AGBE-mediated gene editing. DNA strand with target As and Cs (purple letters) at a locus targeted by a sgRNA (green) is bound by a nCas9(D10A) (blue), which cleaves the non-edited strand. Adenine deamination by an adenine deaminase (yellow) convers the single-strand target A to I (blue letter), which is read as guanosine (G) by polymerase. Following DNA replication or repair, the original A:T base pair is replaced with a G:C base pair at the target site. Cytidine deamination by a cytidine deaminase (pink) convers the single-strand target C to U (blue letter). The resulting G:U heteroduplex induces three possible pathways for DNA repair. Once the U-base is recognized and excised by uracil-DNA glycosylase (UNG), an apurinic/apyrimidinic (AP) site forms (by UNG [purple circle]). (1) The AP site generates a nick by AP lyase or spontaneous lysis at target site and forms a DSB with another nick generated by nCas9(D10A) at a nearby location, which leads to generate indels through NHEJ (left). Or (2) the AP site initiates base repair process, which leads to C-to-N (N = A/T/C/G) conversion, including the C:G starting state (middle). (3) When excision of U-base is inhibited, the G:U mismatch is processed by mismatch repair (MMR), which preferentially repairs the nicked non-edited strand using the deaminated strand as template (the newly synthesized strand is grey). The resulting G:U heteroduplex can be permanently converted to an A:T base pair following DNA replication or DNA repair (right). Editing window of A-to-I: position 4–8, counting the PAM as position 21–23. Editing window of C-to-U: position 3–13.
To validate its practical application, we employed the AGBE to generate a saturated mutant population with base substitutions and indels in the human diphtheria toxin (DT) receptor (hDTR) gene that resists DT in human embryonic kidney 293 (HEK293) cells. Through amplicon deep sequencing, 59,269 mutant variants with different DT sensitivities were identified using 20 functional sgRNAs targeting the hDTR gene.
MATERIALS AND METHODS
Vector construction
The adenosine deaminases, cytidine deaminases, nCas9(D10A) and Escherichia coli-UNG proteins of miniAGBE/AGBEs constructs were amplified from pCMV_ABEmax (#112095), ABE8e (#138489), pCMV-hA3A-BE3 (#113410), CGBE1 (#140252) and miniCGBE1 (#140263), respectively. All of them were cloned into a mammalian expression vector backbone under the control of a CMV promoter by ClonExpress MultiS One Step Cloning Kit (Vazyme, C113). All miniAGBE-puro-EGFP were encoded for co-translational expression of miniAGBEs and PGK-puro-T2A-EGFP. PCR were performed using Q5 High-Fidelity 2X Master Mix (NEB, M0492S). All sgRNA constructs used in this study were designed in accordance with G-N19-NGG rule and cloned into the BpiI-digested (ThermoFisher, FD1014) acceptor vector under a U6 promoter (#48962) by ligating reaction with Solution I (Takara, 6022) for 1 h at 16 °C. All newly constructed vectors were confirmed by Sanger sequencing. All of the primer sets used in this study are listed in Supplementary Table S1 and were synthesized by AZENTA and IGEbio.
Cell culture and electro-transfections
HEK293 cells were cultured and passaged in Dulbecco's modified Eagle's medium (DMEM; HyClone, SH30243.01) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco, 10270-106), and porcine fetal fibroblasts (PFFs) were cultured in DMEM supplemented with 15% (v/v) FBS (Gibco, 10099-141C) and 1 × penicillin/streptomycin (Gibco, 15140122) with 1% nonessential amino acids (Gibco, 11140050), 2 mM GlutaMAX (Gibco, 35050061) and 1 mM sodium pyruvate (Gibco, 11360070). The culture dishes of HEK293 cells were incubated, maintained and cultured at 37.5°C with 5% CO2, and PFFs were maintained at 38.5°C with 5% CO2. Before electro-transfection, cells were digested with 0.25% trypsin (Gibco, 25200-052) and collected. Collected cells were resuspended in 100 μl suspension buffer and co-electro-transfected with 2 μg of sgRNA-expressing vector and 6 μg of base editor-expressing vector at 1150 V, 20 ms and 2 pulse (HEK293 cells) or 1350 V, 30 ms and 1 pulse (PFFs) by using the Neon™ transfection system (Life Technology). At 72 h post-electro-transfection, cells were collected and then used as a template for subsequent PCR to detect efficiencies of gene editing in genome.
Diphtheria toxin (DT) and puromycin treatments in vitro
Transfected HEK293 cells for human diphtheria toxin receptor (hDTR) mutagenesis were selected with 20 ng/ml DT (List Labs, #150) from day 3 after electro-transfections. Cell clones mutated in hDTR-sgRNA-1 or hDTR-sgRNA-2 targeted regions were cultured with DT-supplemented growth medium from day 1 after cell sorting (day 0 refers to the day of cell sorting). Transfected PFFs were selected with 300 ng/ml puromycin-containing (MPBIO, 219453925) medium for 48 h after electro-transfections of 12 h. Both DT and puromycin-supplemented growth medium were exchanged daily until negative control cells died completely.
Transcription of RNA in vitro
Vector pcDNA3.1-miniAGBE-4 containing a T7 promoter in front of the coding sequence was linearized by restriction endonuclease MssI (ThermoFisher, FD1344) and then used as transcription template of miniAGBE mRNA. The miniAGBE-4 mRNA was synthesized using HiScribe™ T7 ARCA mRNA Kit (with tailing) (NEB, E2060S). By using specific forward primers, we amplified sgRNAs with a U6 promoter with each pT7-pBRCA2-F, pT7-pRAG2-F, and pT7-pPPARγ-F to introduce a T7 promoter sequences (TAATACGACTCACTATAGGG) and construct pT7 driving expression sgRNA scaffolds together with consistent reverse primer T7-template-R (GGGTCTAGAAAAAAAGCACCGAC). The PCR products were then transcribed using HiScribe™ T7 Quick High Yield RNA Synthesis Kit (NEB, E2040S). Both mRNA and sgRNAs were purified with RNeasy MiniElute Cleanup kit (Qiagen, 74204) following the manufacturer's instructions. The purified mRNA of miniAGBE and sgRNA were diluted to 1000 ng/μl and 300 ng/μl with RNase-free water, respectively, and stored at −80 °C. They were mixed in a 3:1 ratio before microinjection at a final concentration of 150 and 50 ng/μl, respectively. The primers used for amplifying template of sgRNAs are listed in Supplementary Table S1.
Pig embryo collection, microinjection and identification
In theory, porcine zygotes from Large White pigs were collected after insemination and transferred into manipulation medium. Given that the porcine oocytes are easily available from a local slaughterhouse, in vitro-activated porcine parthenogenetic (PA) embryos were used to evaluate the editing efficiency in pig embryos after injection with a mixture of RNAs. Porcine oocyte collection, in vitro maturation, and parthenogenetic activation were conducted as previous described (27,28). In brief, a mixture of miniAGBE mRNA (150 ng/μl) and sgRNA (50 ng/μl) was microinjected into the cytoplasm of porcine PA embryos. After 6-day in vitro culture, blastocysts were collected for genotyping by PCR and Sanger sequencing.
Genomic DNA extraction and genotyping
HEK293 cells, PFFs, and porcine blastocysts were collected in 10 μl (HEK293 cells and PFFs) and 6 μl (porcine blastocysts) of lysis buffer (0.45% NP-40 plus 0.6% proteinase K), respectively, and lysed at 56°C for 60 min, followed by a 96 °C enzyme inactivation step for 10 min. The cell lysates were used as PCR templates for editing efficiencies analysis. With the above purpose, specific primer pairs of target region and 2 μl cell lysate were applied to amplify DNA fragments including the edited site. The PCR products were then directly sent for Sanger sequencing or cloned into T vector for sequencing.
TA clone sequencing
TA clone sequencing here was mainly used to confirm genotypes of specific cell clones with hDTR mutation. The target sequence was first generated from the genome of cell clone by PCR with the 2× Phanta Max Master mix (Vazyme, P515-01) and specific primers. Then, an adenine deoxyribonucleotide was added to the PCR products with Taq polymerase at 72 °C for 10 min (10 μl reaction mix, consisting of 5 μl 2× Rapid Taq Master Mix [Vazyme, P222-03] and 5 μl PCR product). The final product was cloned into pMD™18-T Vector (Takara, 6011) with Solution I at 16 °C for 2 h (10 μl reaction mix, consisting of 4.5 μl final PCR product, 0.5 μl pMD™18-T Vector, and 5 μl Solution I). Ligation product was transfected to 50 μl competent cells and cultured at 37°C until proper single bacteria colony occurred. Ten single bacterial colonies were sent for vector extraction and then Sanger sequencing.
Editing efficiency detection and analysis by Sanger sequencing
HEK293 cells and PFFs were electro-transfected with indicated AGBEs and sgRNAs. After 72 h, HEK293 cells were collected without sorting or selecting, and PFFs were collected with puromycin selecting for subsequent PCR amplification. Editing efficiency was detected by Sanger sequencing, comparing with control groups. The Sanger sequencing results of AGBEs were analysed by EditR (https://moriaritylab.shinyapps.io/editr_v10/) (29) for quantification.
Whole-genome sequencing (WGS) and bioinformatics analysis
Five groups with electro-transfected HEK293 cells were used for WGS analysis: cells electro-transfected with miniAGBE-4 with a sgRNA targeting hABE site 1 (AGBE-hABE site 1), or a sgRNA targeting hABE site 7 (AGBE-hABE site 7), or a non-targeting sgRNA (AGBE-NT); cells electro-transfected with miniAGBE-4 alone or without transfection of miniAGBE-4 and sgRNA were used as negative control and WT control, respectively. Triplicate trails were conducted for all the five groups. About 106 cells (EGFP+) of each sample were collected by fluorescence-activated cell sorting (∼48–72 h post electro-transfection) and genomic DNA were extracted by Cell Genome Extraction kit (Tiangen Biotech, DP304-03). The WGS libraries were prepared by HaploX Biotechnology (Jiangxi, China) and then sequenced at mean coverages of 30× by Illumina Nova 6000. The raw data were filtered and trimmed using fastp (v0.20.1) with the base quality value ≥15 (-q 15). The qualified short reads were mapped to human genome (GCF_000001405.39 from NCBI) using BWA (v 0.7.15-r1140) MEM algorithm. After the initial alignment, Samtools (v1.3.1) was used to process aligned BAM files. Picard (v2.24.2) was run to remove duplicate reads in the mapped BAM files. The genome-wide variants were called using HaplotypeCaller (GATK v4.2.0.0), and known variants in dbSNP version 146 were used during base quality recalibration. Variants pass the filter expression ‘QUAL ≥ 20 & DP ≥ 30 & QD ≥ 2 & MQ ≥ 20 & FS ≤ 6 & SOR ≤ 2’ were considered as high-confidence variant calls. Variant annotation and further filtration were conducted using ANNOVAR (version 2019Oct24) and whole-genome databases avsnp150. Variants annotated to known sites were eliminated in the subsequent analysis. These final, high-confidence variant calls for each treated sample were used for downstream analysis. Genomic tracks of single-nucleotide variants (SNVs) and indels were drawn using Circlize (version 0.4.4), and ggplot2 (3.3.0) was used for other customized visualizations.
Transcriptome sequencing (RNA-seq) and bioinformatics analysis
For transcriptome analysis, ∼106 cells (EGFP+) of each sample were collected and used for RNA extraction (Magen, R4012-03). The RNA-seq libraries were constructed by HaploX Biotechnology (Jiangxi, China) according to the standard protocol. High-throughput transcriptome sequencing was carried out using Illumina Nova 6000 according to the manufacturer's instructions. As the whole-genome sequencing data analysis described above with small modification. Simply, raw data were filtered and trimmed using fastp (v0.20.1) with the base quality value ≥15 (-q 15). The qualified short reads were mapped to human genome (GCF_000001405.39 from NCBI) using STAR (v2.7.0f_0328). Samtools (v1.3.1) and Picard (v2.24.2) were used to process aligned BAM files and remove duplicate reads. The transcriptome-wide variants were called using HaplotypeCaller (GATK v4.2.0.0) with the additional parameters ‘–min-pruning 0, –dont-use-soft-clipped-bases, –linked-de-bruijn-graph and –recover-all-dangling-branches’. These final, high-confidence variant calls were used for downstream analysis.
Amplicon deep sequencing and data analysis
The target sites were amplified with site-specific primer pairs for 30 cycles with 98°C 10 s, 58°C 30 s, 72°C 8 s by Q5 DNA polymerases (NEB, M0492S) from cell lysate of the electro-transfected cells or cell clone-derived from hDTR mutations. The PCR products were then used as a template for subsequent amplification with different index primers of different samples at 98°C 10 s, 58°C 30 s, 72°C 8 s for 12 cycles by Q5 DNA polymerases. PCR products of all samples were gel-purified using HiPure Gel Pure DNA Mini Kit (Magen, D1001-03). The concentration of purified products was quantified with an IPure Qubit dsDNA HS assay kit (IGE Biotech, IGE2019052901). Then, equivalent amounts of purified products were mixed together to make a DNA library, which was sent to Annoroad Gene Technology Co., Ltd, Beijing, China for amplicon deep sequencing by using a NovaSeq platform. The protospacer sequences in reads were investigated to identify different point mutation types and indels. The presented ratio was calculated by comparing individual reads to the whole reads. The amplicons were sequenced in three of the repeated assays for each target site. Amplicon reads with a quality score of <30 were filtered. Primer sets used for amplicon deep sequencing are listed in Supplementary Table S1.
Plotting of cell growth curve
To determine whether mutations introduced into hDTR by miniAGBE-4 confer DT-resistance to HEK293 cells, we quantified cell numbers of 6 hDTR-mutated cell clones and HEK293 wildtype (WT) cell to evaluate cell growth rate. Each 2000 cells were sorted using BD FACSAria II and seeded per well of a 24-well plate (Corning). DT-supplemented growth medium was exchange 24 h later after sorting. Cell number of each cell clone was recorded every 24 h for 7 days by manual counting with blood cell counting board (Qiu Jing). Each clone was quantified for three duplicate wells and each well was counted by three biological replicates and averaged every day.
RT-PCR
Total RNA was extracted from cell clones using RaPure Total RNA Micro Kit (Magen, R4012-03) according to manufacturer's instructions. cDNA synthesis was performed using HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, R312-02). Sanger sequencing after PCR was performed with primers as follows, F: 5′-GTGCCTCCCTCCTCCCCAT-3′; R: 5′-CACAAAGCCAGTTTCCCCA-3′.
Statistical analysis
Data were statistically analysed by using Excel and GraphPad Prism v.8.0. Editing frequencies of different cytosines and adenines in the spacer sequence were analysed on an online tool, EditR 1.0.10 (https://moriaritylab.shinyapps.io/editr_v10/) (29).The simultaneous base editing frequencies and indels were calculated from data of amplicon deep sequencing. All the numerical values of target sites were presented as means ± s.d.. A statistical comparison adjustment was performed using two tailed Student's t-test. P < 0.05 was considered as a significant difference between treatment groups.
RESULTS
Construction and optimization of AGBEs
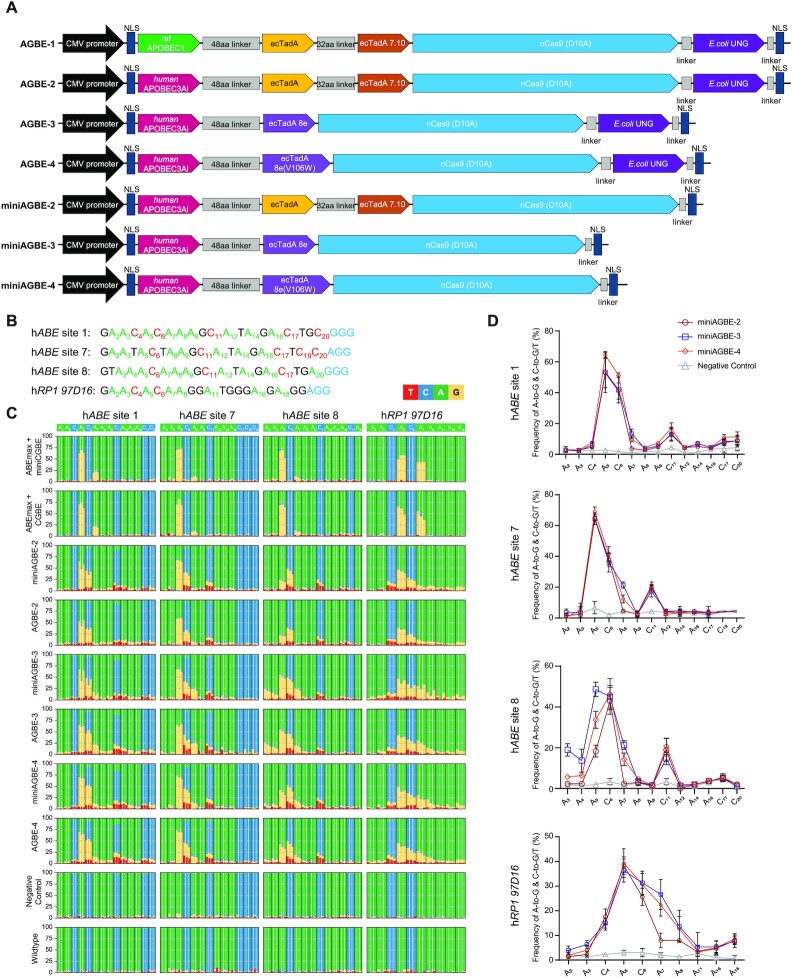
AGBEs are composed of two deaminases (cytidine deaminase and adenine deaminase), a nCas9(D10A) and an E. coli-derived UNG (eUNG). Similar to previously reported CGBEs (22,23), the first version of AGBE (AGBE-1) was constructed by fusing a rat-derived cytidine deaminase variant, rat APOBEC1(R33A), to the N-terminus of ABEmax with an eUNG at the C-terminus of ABEmax (Figure 2A). Given that human APOBEC3A-conjugated BEs can mediate higher C-to-T base editing than rat APOBEC1 (30–34), we initially attempted to create the second AGBE version (AGBE-2) by simply replacing rat APOBEC1(R33A) with human APOBEC3A (hAPOBEC3A) (Figure 2A). However, during the process of constructing the vectors, we were unable to obtain the correct vectors with the hAPOBEC3A-nCas9(D10A) fusion gene from E. coli competent cells, indicating that this fusion protein could be toxic to bacteria (Supplementary Figure S1A). We speculated that as a deaminase with increased efficiency, hAPOBEC3A induced more extensive deamination of the cytosine (C) base in the prokaryotic genome, reducing the stability of the genome in the absence of the UGI domain. To overcome this problem, an artificial intron was inserted into the coding sequence (CDS) of hAPOBEC3A to prevent the expression of functional APOBEC3A protein due to the lack of an endogenous mRNA splicing machinery in prokaryotic cells (Supplementary Figure S1B) (35). Using this modified APOBEC3A, named APOBEC3Ai, the functional mRNAs of hAPOBEC3A deaminase could be formed since eukaryotic cells possess endogenous mRNA splicing machinery.
Figure 2.
Base editing activities of AGBEs with different architectures in HEK293 cells. (A) Architectures of miniAGBEs and AGBEs. aa, amino acid; NLS, nuclear localization signal. (B) Protospacers and PAM (blue) sequences of four human genomic loci studied by AGBEs, with target As (green) and Cs (red) in (C). (C) Bar plots shows on-target As (green) and Cs (blue) base editing frequencies induced by various AGBEs with four sgRNAs targeting genomic loci in HEK293 cells. Editing frequencies of three independent replicates at each base are display side-by-side. (D) Comparison of A-to-G and C-to-G/C-to-T editing frequencies induced by three miniAGBEs (The indistinctive conversion of C-to-A are not shown). Values and error bars indicate the mean ± s.d. of three independent replicates. Subscript number indicates position of the base in the protospacer, counting the PAM as position 21–23. HEK293 cells electro-transfected with sgRNA only served as negative control group, and cells electro-transfected with resuspension buffer only served as WT group. The editing efficiency in (C and D) is analysed by EditR for quantification.
In the preliminary experiment, we co-electro-transfected AGBE-1 and AGBE-2 together with sgRNAs targeting human endogenous genes into HEK293 cells. Sanger and amplicon deep sequencing showed that AGBE-2 produced more abundant gene editing outcomes, whereas AGBE-1 achieved mainly A-to-G base substitution (Supplementary Figures S2A–E).
Based on AGBE-2, we further constructed another two AGBE versions by replacing the ecTadA-TadA*7.10 heterodimer with the monomers ecTadA8e (AGBE-3) and ecTadA8e(V106W) (AGBE-4) (Figure 2A), as both ABE8e and ABE8e(V106W) exhibited substantially greater editing efficiency than ABEmax (36–38).
Previous reports have suggested that the existence of a UNG domain (22) or a UNG ortholog (UdgX) (25) in CGBEs results in comparable or only slightly altered editing frequencies. Hence, to reduce the size of AGBE, three smaller AGBEs, namely, miniAGBE-2, miniAGBE-3 and miniAGBE-4, were constructed by removing of the eUNG domain (Figure 2A).
Validation and comparison of the editing activity of AGBEs in HEK293 cells
To evaluate the base editing activities of the AGBEs on endogenous genes, seven sgRNAs (hABE site 1, hABE site 7, hABE site 8, hRP1 97D16, hHEK4 OT2, hRNF2 site 1 and hPPP1R12C site 6) targeting human genomic genes were designed according to the CGBEs (22,23) (Figure 2B and Supplementary Figure S3A). Each sgRNA was co-electro-transfected into HEK293 cells along with each of the six AGBEs (AGBE-2–AGBE-4 and miniAGBE-2–miniAGBE-4). Cells electro-transfected with ABEmax, CGBE (or miniCGBE) (22) and sgRNA-expressing vectors or sgRNA-expressing vectors only were used as controls. Three days post-transduction, the cells were collected, and whole genomic DNA was extracted to determine the editing frequencies and editing windows. Sanger sequencing of the PCR products surrounding the target sites confirmed that all six AGBEs produced simultaneous C-to-G/T/A and A-to-G conversions; in contrast, when co-electro-transfected alone, ABEmax and CGBE/miniCGBE only achieved one type of base substitution at the tested sites, i.e. A-to-G or C-to-G/T/A substitution. The base editing efficiencies were then quantified using a novel base editing quantification software program, EditR (29). For A-to-G conversion, the editing windows of the AGBEs were consistent with that of ABEmax (39) (from positions 4–8, counting the protospacer-adjacent motif [PAM] as positions 21–23) at most of the target sites (Figures 2C, D and Supplementary Figures S3B, C and S4), whereas the editing efficiencies of the AGBEs with the ecTadA-TadA*7.10 heterodimer were markedly lower than those of ABEmax, which might have been obtained because the function of the ABE was disturbed by the competition with CGBE (17). However, when the ecTadA-TadA*7.10 heterodimer was replaced with ecTadA8e or ecTadA8e(V106W), the AGBE variants had A-to-G editing efficiencies comparable to that of ABE alone (Figures 2C, D and Supplementary Figures S3B, C and S5). For cytosine base editing, the editing window expanded from positions 4–8 to 3–13. The C-to-G, C-to-T and C-to-A conversions were generated at all sites, and different efficiencies were detected at different sites.
The PCR products were then subjected to amplicon deep sequencing to determine the abundances of mutation patterns and whether C-to-G/T/A and A-to-G base editing occurred simultaneously at the same DNA strand. The results revealed that hAPOBEC3Ai-derived AGBE systems induced simultaneous A-to-G and C-to-G (0.01–32.46%), A-to-G and C-to-T (0.18–23.75%), or A-to-G and C-to-A (0.01–2.59%) conversions at the same DNA strand, while concurrent A and C editing was almost undetectable in cells co-electro-transfected with single base editors (A-to-G and C-to-G: 0.13–0.90%; A-to-G and C-to-T: 0.07–0.58%; A-to-G and C-to-A: 0.01–0.30%) (Figure 3A, Supplementary Figure S6A and Supplementary Table S2). In addition, three or more simultaneous types of base substitutions (such as simultaneous C-to-A, C-to-G, and A-to-G; C-to-A, C-to-T, and A-to-G; C-to-T, C-to-G, and A-to-G; and C-to-A, C-to-G, C-to-T, and A-to-G substitutions) were also found with efficiencies ranging from 0.01% to 10.35%.
Figure 3.
Comparison of gene editing products induced by various AGBEs at four target sites in HEK293 cells. (A) Comparison of base editing products distribution (top) and indel frequencies (below) among four human genomic loci in HEK293 cells treated with AGBEs and the corresponding sgRNA, or in control groups. Editing frequencies reflect sequencing reads that contain base editing only and do not contain indels among all treated cells, without sorting. (B–E) Allele frequencies of DNA on-target editing within four human genomic loci by miniAGBE-2, miniAGBE-3 and miniAGBE-4, respectively (n = 1) in HEK293 cells. The values in right represent frequencies and reads of mutation alleles. Data are taken from the first replicate obtained for each sgRNA from the on-target experiment shown in Figure 2C. HEK293 cells electro-transfected with sgRNA only served as negative control group, and cells electro-transfected with resuspension buffer only served as WT group.
In addition to base editing, the indel efficiencies of the hAPOBEC3Ai-derived AGBE systems in the seven tested loci (AGBE-2: 4.10–19.73%; AGBE-3: 7.21–35.92%; AGBE-4: 6.95–40.89%; miniAGBE-2: 5.64–33.42%; miniAGBE-3: 5.41–32.04% and miniAGBE-4: 5.54–43.01%) were significantly higher than those of ABEmax + miniCGBE (0.29–2.44%) and ABEmax + CGBE (0.36–2.77%) (Figure 3A, Supplementary Figure S6A and Supplementary Table S2).
We further analysed the details of the outcomes yielded by AGBEs in HEK293 cells. Due to the random editing of Cs and the expanded editing windows within the protospacer, hAPOBEC3Ai-based AGBEs yielded diverse mutation types with different editing events, and most of these were desired (Supplementary Figure S7). Using hABE site 1 as an example, AGBE-2, AGBE-3, AGBE-4, miniAGBE-2, miniAGBE-3 and miniAGBE-4 induced 1,603, 2,387, 2,592, 1,752, 2,124 and 2,234 types of mutational SNVs with the same sgRNA, respectively, while only 836 and 926 mutation patterns were observed in the ABEmax + miniCGBE and ABEmax + CGBE groups, respectively (Supplementary Table S3). Thus, AGBEs can be used to improve the gene editing diversity for the establishment of saturated mutant populations.
We subsequently evaluated whether eUNG was indispensable for the architecture of AGBEs. Sanger sequencing and amplicon deep sequencing of these seven target sites showed no significant difference in the editing frequency, editing window or number of mutation types after the conjugation of eUNG. The smaller size of the miniAGBEs (Δ684 bp) (22) is an advantage for achieving higher vector transfection efficiency; thus, we used the miniAGBEs in the subsequent experiments.
The top 10 patterns of genome modification mediated by miniAGBE-2–miniAGBE-4 are separately listed in Figures 3B–E and Supplementary Figures S6B–D. For hABE site 1 and hABE site 7, the most frequent gene editing pattern was simultaneous A5-to-G and C6-to-G editing, and the efficiencies ranged from 14.73% to 27.76%. In addition, other patterns of simultaneous heterogeneous base editing (such as A5-to-G and C6-to-T; A5-to-G, C6-to-G and C11-to-T; A5-to-G and C11-to-T; A5-to-G, C6-to-G and C11-to-G; A5-to-G, C6-to-G and A7-to-G; A5-to-G, C6-to-T and A7-to-G) were also found with efficiencies ranging from 0.48% to 7.88%. For hABE site 8 and hRP1 97D16, simultaneous heterogeneous base editing at the same DNA strand was also observed with somewhat low efficiencies ranging from 0.74% to 10.18%. However, for hHEK4 OT2, hRNF2 site 1 and hPPP1R12C site 6, simultaneous heterogeneous base substitutions at the same DNA strand were almost undetectable with efficiencies lower than 1%. All the above-described results suggest that the AGBE system provides a flexible tool for the efficient generation of multiple homogeneous base editing, heterogeneous base editing, and indels.
The miniAGBEs edit efficiently in PFFs and porcine embryos
We subsequently verified whether the miniAGBEs could efficiently induce multiple patterns of genome modifications in primary somatic cells and embryos. PFFs and embryos were selected for further verification. Three sgRNAs targeting porcine BRCA2, PPARγ and RAG2 were designed. Individual sgRNAs with miniAGBE-2–miniAGBE-4-expressing vectors were co-electro-transfected into PFFs. After 3 days of puromycin screening, the surviving PFFs were collected and analysed by Sanger sequencing and amplicon deep sequencing. Sanger sequencing revealed that miniAGBE-3 and miniAGBE-4 induced comparable A-to-G, C-to-G and C-to-T base substitutions in primary somatic cells, while miniAGBE-2 exhibited substantially decreased base editing at all three tested loci (Figure 4A). Instead of simultaneous A-to-G and C-to-G conversions in HEK293 cells, the miniAGBEs mainly introduced simultaneous A-to-G and C-to-T conversions in PFFs (pBRCA2: 11.05%, pPPARγ: 36.29%, pRAG2: 13.20%) (Figure 4B, Supplementary Figures S8A-C and Supplementary Table S4). The analysis of deep sequencing reads revealed that the miniAGBEs generated many more abundant types of base-edited mutants (pBRCA2: 1,761 ± 76; pPPARγ: 1,429 ± 89; pRAG2: 1,959 ± 235) than WT PFFs (pBRCA2: 674 ± 29; pPPARγ: 341 ± 86; pRAG2: 529 ± 51) (Supplementary Figure S8D and Supplementary Table S5). Moreover, the miniAGBEs induced a higher indel frequency in PFFs than in HEK293 cells, which ranged from 17.71% to 71.82%, and these differences might have been related to the cell types and gene loci. miniAGBE-3 showed the highest frequency of A and C co-conversion (ranging from 4.18% in pRAG2 to 12.72% in pPPARγ) (Figure 4B and Supplementary Table S4) across all tested sites, followed by miniAGBE-4 (ranging from 3.96% in pRAG2 to 10.99% in pPPARγ). Both of them shared comparable numbers of mutation types (295 ± 89 and 323 ± 115, respectively) (Supplementary Figure S8D and Supplementary Table S5). Given that the adenine deaminase mutant ecTadA8e(V106W) in miniAGBE-4 can minimize possible off-target RNA editing, we used miniAGBE-4 in the subsequent trials.
Figure 4.
Gene editing activities of miniAGBEs at three target sites in PFFs and porcine embryos. (A) Comparison of A-to-G, C-to-G and C-to-T editing frequencies at 3 endogenous porcine genomic loci by miniAGBE-2, miniAGBE-3, and miniAGBE-4, respectively (n = 3) in PFFs (The indistinctive conversion of C-to-A are not shown). (B) Comparison of base editing products distribution (top) and indel frequencies (below) among edited porcine genomic loci in PFFs treated with miniAGBE-2, miniAGBE-3 and miniAGBE-4 and the corresponding sgRNA, or in control groups. Values and error bars indicate the mean ± s.d. of three independent replicates. Editing frequencies reflect sequencing reads that contain base editing only and do not contain indels among all treated cells, with puromycin selecting. PFFs electro-transfected with resuspension buffer only served as WT control. ns, no significant difference (P> 0.05). (C) Summary of porcine embryo development with miniAGBE-4. Embryo injected with sterile water served as WT control. ns, no significant difference (P> 0.05). (D) Scatter plots shows base editing frequencies in porcine embryos by co-delivering miniAGBE-4 mRNA (150 ng/μl) with each three sgRNAs (50 ng/μl) in (A) via micro-injection. Each dot indicates an individual embryo. Data are presented as mean ± s.d., and the bold lines are represented the mean of base editing frequencies.
Subsequently, to validate the effectiveness of miniAGBE-4 in mammalian embryos in vitro, porcine parthenogenetically activated (PA) oocytes were collected and injected with a mixture of miniAGBE-4 mRNA and the three sgRNAs that were effective in PFFs. The injected embryos were cultured for 6 days after parthenogenetic activation until blastocyst formation. No significant difference in the blastocyst rate was found between the embryos injected with RNA (48.2%, 45.1%, and 50.7%) and the control embryos (53.3%) (Figure 4C and Supplementary Table S6). The blastocysts were collected to detect the editing frequency. Sanger sequencing revealed that miniAGBE-4 exhibited high base conversion efficiency (A-to-G up to 73% at pPPARγ, C-to-G up to 22% at pBRCA2, and C-to-T up to 52% at pPPARγ) (Figure 4D).
DNA and RNA off-target effects analysis for miniAGBE-4
Base editors have been reported to exhibit potential DNA and RNA off-target effects, raising safety concerns regarding the AGBE system (40,41,42,43). Thus, we performed both sgRNA-dependent and deaminase-dependent off-target analyses to characterize the potential off-target effects of miniAGBE-4 in HEK293 cells. The miniAGBE-4 was electro-transfected into cells with a sgRNA targeting hABE site 1 (AGBE-hABE site 1), or a sgRNA targeting hABE site 7 (AGBE-hABE site 7), or a non-targeting sgRNA (AGBE-NT). Cells electro-transfected with miniAGBE-4 alone or without transfection of miniAGBE-4 or sgRNA were used as negative control and wildtype control, respectively. Triplicate trials were conducted for all five groups.
First, we performed whole-genome sequencing (WGS) to assess both sgRNA-dependent and deaminase-dependent DNA off-target activities of miniAGBE-4. The TOP20 potential off-target sites of each sgRNA were selected by Cas-OFFinder prediction (44) and analysed by searching the WGS dataset for all similar target sites that contained up to a 4-nucleotide mismatch. The sites we scanned did not reveal any notable sequence alterations in comparison to the WT one (Supplementary Table S7). For the deaminase-dependent off-target analysis, we retrieved the de novo SNVs and indels from the WGS dataset of all groups. As shown in Supplementary Figures S9A–E and Supplementary Tables S8-S9, miniAGBE-4 did not induce detectable deaminase-dependent off-target editing in terms of the number, distribution or proportion of SNVs and indels at the genome-wide level. The overlap scope of SNV numbers displayed in the Venn diagram among different groups was not distinct, indicating that they might be derived from natural random mutations (Supplementary Figure S9F and Supplementary Table S10). To examine transcriptome-wide off-target effects, we performed WGS-matched side-by-side RNA sequencing (RNA-seq) of total RNA of miniAGBE-4-expressing cells. Given that miniAGBE-4 was fused with the ecTadA8e V106W mutant and hAPOBEC3Ai, we focused on the A-to-I substitution and C-to-G/U/A substitution induced by the two deaminases, respectively. As shown in Supplementary Figure S10 and Supplementary Tables S11–S12, the number and proportion of de novo SNVs and indels in the RNA-seq dataset were not elevated in the groups with cells expressing miniAGBE-4 with sgRNAs, which were comparable to the control groups. Moreover, the off-target RNA SNVs were found in both coding and non-coding sequences, which was consistent with previous reports (40,41). The results above indicated that miniAGBE-4 enabled the efficient on-target editing and did not induce dramatically different DNA or RNA off-target effects compared with the wildtype HEK293 cells.
Installation of DT-resistant mutations in human cells with miniAGBE-4
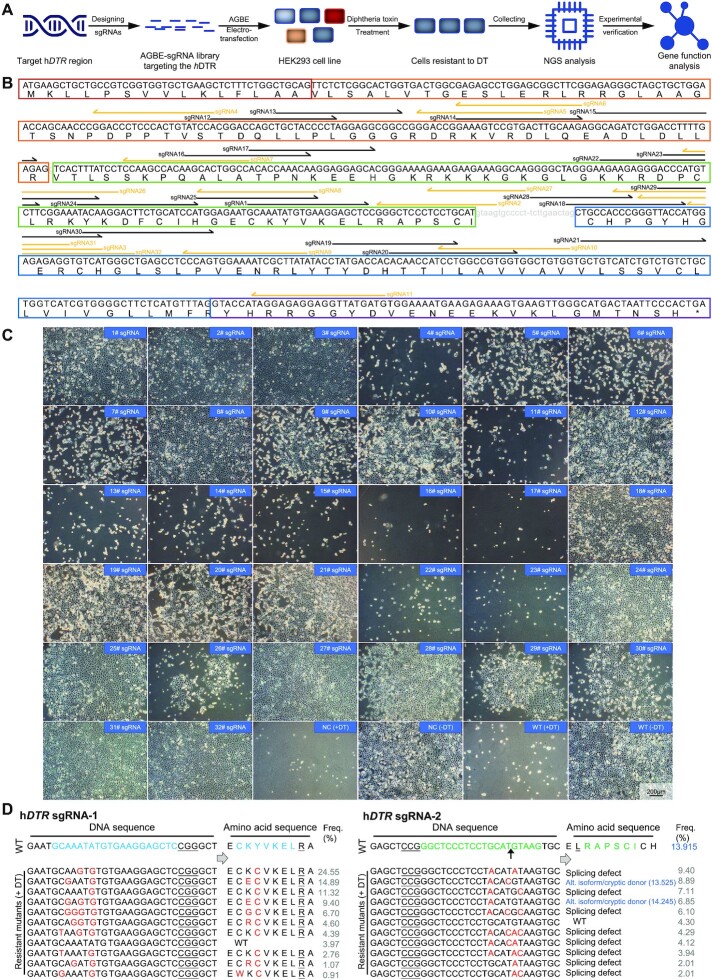
To demonstrate the utility of AGBEs in screening for functional mutations through saturated mutagenesis, we applied miniAGBE-4 to install loss-of-function mutations in hDTR. If an encoding genetic variant that rendered resistance to DT was created, the human cell would survive the screening and proliferation to form a population (45) (Figure 5A). We designed a library that included 32 sgRNAs with As and Cs in the optimal editing window. Sixteen of these sgRNAs covered the critical region (amino acids Asp106-Ser147) of the human epidermal growth factor (EGF)-like DT binding domain, which acted as the functional DT receptor, while others covered some non-conserved amino acids between mouse and human DTRs (Figure 5B and Supplementary Figure S11A) (46). Each sgRNA was transiently co-electro-transfected into HEK293 cells with miniAGBE-4. Cells electro-transfected with miniAGBE-4 only or with resuspension buffer only were used as negative controls and WT control. Two days later, we observed distinct gene editing at 28 out of 32 sgRNAs (87.50%). After treatment of cells with DT, we were able to determine whether hDTR was inactivated based on the survival of the cells. After 5 days of DT screening, 20 groups (20/28) with sgRNAs targeting the hDTR locus exhibited robust resistance to DT, while the groups of cells electro-transfected with other combinations of vectors, as well as the control cells, did not survive after treatment (Figure 5C). Conversely, cells electro-transfected with miniAGBE-4 and the hDTR sgRNA library maintained a growth rate similar to that of the WT cells in normal medium (Supplementary Figure S11B). We selected cells with mutations introduced by miniAGBE-4 and the 20 sgRNAs for further analysis. Amplicon deep sequencing was performed to reveal the differences in the genotypes of cells cultured in DT-supplemented medium and normal medium. The rate of knockout mutation reads was increased in the cells with hDTR mutations under DT selection (Supplementary Figures S11C, S12A and S13).
Figure 5.
Installation of diphtheria toxin (DT) resistance mutations in hDTR by miniAGBE-4 in HEK293 cells. (A) Workflow of procedure for mutating the hDTR under DT-based selection via miniAGBE-4 in HEK293 cells. NGS, next generation sequencing. (B) Schematic shows the target sites of AGBE-based sgRNA library for hDTR mutation screening. Coloured boxes indicate boundaries of exon region. Gray lowercase letters represent intron sequences. Black (forward) and yellow (reversed) half arrows targeting the below letters indicate protospacer sequences and direction of individual sgRNA. (C) Images of HEK293 cells with DT treatment for 5 days after miniAGBE-4 editing with indicated sgRNAs. NC, HEK293 cells electro-transfected with miniAGBE-4 only. WT, HEK293 cells electro-transfected with resuspension buffer only. +DT, cultured in DT-supplemented medium. -DT, cultured in normal medium. The effect of individual sgRNA had been verified by three biological replicates. Scale bar: 200 μm. (D) Frequencies of hDTR mutations in DT-resistant cells after miniAGBE-4 with sgRNA-1 or sgRNA-2 editing. Gray values in right represent frequency of mutations. Mutations in hDTR sgRNA-2 targeted splice site (5′-GT) resulting in RNA alternative splicing events. Dark blue values in sgRNA-2 represent scores of the WT and predicted emerging splice donor sites by http://wangcomputing.com/assp/. The black arrow indicates boundary of exon and intron sequences. Target DNA sequence and amino acids (blue and green), PAM (underline), mutant sites and amino acids (red). Splicing defect means the splice site is destroyed. Alt. isoform/cryptic donor means the alternative isoform or cryptic splice site may be activated.
A total of 59,269 types of base-edited alleles induced by the 20 sgRNAs were achieved after treatment with DT (Supplementary Figure S12B and Supplementary Table S13). Among them, 54.23% (32,140/59,269) contained concurrent A-to-G (T-to-C) and C-to-G/T/A (G-to-C/A/T) conversions (Supplementary Figure S12A and Supplementary Tables S13-S14). Based on our analysis of the TOP10 mutations, Cys121, Tyr123, Glu141 and Thr160 appeared to play a key role in conferring DT insensitivity to human cells because most amino acid substitutions were induced in these residues: 80.59% were Y123C mutations for sgRNA-1, 25.69% were C121Y/Y123H synchronous mutations for sgRNA-8, and 5.69% were T160I mutations for sgRNA-19 (Figure 5D and Supplementary Figures S11C and S13).
A recent study developed a new approach called BARBEK, which leverages CBEs and iBARed sgRNAs for the genome-scale interrogation of gene functions via perturbation of gene splice sites (13). Inspired by this concept, 4 out of 32 sgRNAs (sgRNA-2, sgRNA-18, sgRNA-27 and sgRNA-28) targeting the splice donor site (5′-GT) and splice acceptor site (5′-AG) were designed to disrupt splicing of hDTR precursor-mRNAs (Figure 5B). Amplicon deep sequencing indicated that most of the conserved splice site motifs at these four targets were mutated. These mutations might have resulted in altered RNA-splicing events, including splicing defects or activation of a cryptic splice site (47) (based on the scores predicted by an online tool, http://wangcomputing.com/assp/), or in direct disruption of gene function (48,49) (Figure 5D and Supplementary Figures S11C and S13).
Characterization of the functions of hDTR variants generated by miniAGBE-4
We then characterized the functions of the hDTR variants to verify whether this AGBE system could be used to study mutations associated with specific phenotypes. hDTR sgRNA-1 targeting the CDS and hDTR sgRNA-2 targeting the 5′ splice site of intron 3 (Figure 5D), both of which were expected to interfere with hDTR expression, were selected to create variants in cells for functional characterization.
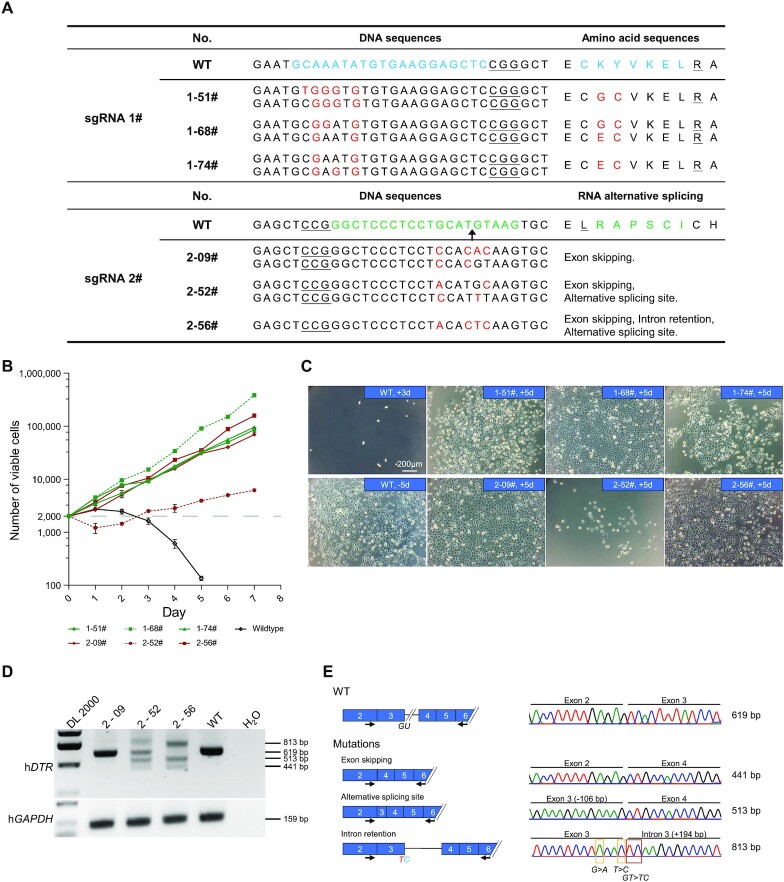
First, we used flow cytometry to isolate single cells from bulk-edited cells and obtained corresponding cell clones after culturing the cells in normal medium for a period of time. Through Sanger sequencing of genomic DNA from the mutant clones, we identified 73 and 76 clones with hDTR mutations from sgRNA-1-derived cells and sgRNA-2-derived cells, respectively. A total of 76.7% (56/73) of sgRNA-1-derived clones had A or C conversions within the protospacer, and 76.3% (58/76) of sgRNA-2-derived clones had T4-to-C or G5-to-A/T conversions at the desired T and G bases of the 5′ splice site (Supplementary Figure S14 and Supplementary Table S15). Subsequently, 6 clones with a variety of editing types were cultured under DT pressure for 7 days. Clone 1–68# exhibited the fastest proliferation rate, followed by Clone 1–51#, Clone 1–74#, Clone 2–09# and Clone 2–56#. The cells of Clone 2–52# were alive but displayed no growth, whereas those of the WT group died after 3 days (Figures 6B, C).
Figure 6.
Characterization of the function of hDTR variants generated by miniAGBE-4. (A) Genotypes and corresponding amino acid substitution (sgRNA-1) or RNA alternative splicing events (sgRNA-2) of hDTR-mutated single-cell clones derived from miniAGBE-4/sgRNA-1 and miniAGBE-4/sgRNA-2 edited HEK293 cells. Target DNA sequence and amino acids (blue and green), PAM (underline), mutant sites and amino acids (red). (B) Cell proliferation in DT-supplemented medium. Wildtype HEK293 cells are almost completely eliminated by DT after 3 days culturing. Values and error bars indicate the mean ± s.d. of three independent replicates. (C) Images of DT-insensitive single-cell clones with DT treatment for 5 days. +3d/+5d, cultured in DT-supplemented medium for 3 or 5 days. -5d, cultured in normal medium for 5 days. Scale bar: 200 μm. (D) RT-PCR analysis of isoforms expression in WT HEK293 cells and single-cell clones derived from miniAGBE-4/sgRNA-2-edited cells. Data are the representative of three independent replicates. DL 2000, DNA marker. (E) Schematic diagram of hDTR mRNA variants in WT and mutated cell clones edited the 5′ splice site of exon 3 in hDTR and details of the sequences of RT-PCR amplicons.
Sanger sequencing indicated that Clone 1–51# and Clone 1–74# were homozygotes with the K122G/Y123C and K122E/Y123C mutations, respectively, and Clone 1–68# was heterozygous with the K122E/Y123C and K122G/Y123C biallelic mutations (Figure 6A and Supplementary Figure S14B). As sgRNA-2 targeted the conserved motif of the 5′-splice site, whether the transcription of hDTR was disrupted in these clones needed to be investigated further. We performed RT-PCR with a forward primer in exon 2 and a reverse primer in exon 6 to examine the product of hDTR splicing. A 619-bp fragment was detected in WT cells, whereas 1, 4, and 3 fragments were observed in Clone 2–09#, Clone 2–52# and Clone 2–56#, respectively (Figure 6D). Sanger sequencing confirmed that skipping of exon 3, retention of intron 3, and activation of a cryptic splice site of exon 3 occurred in these cell clones (Figure 6E). The above-described results indicate that miniAGBE-4 can generate a range of variants with different DT sensitivities by editing the CDSs or splice sites.
DISCUSSION
In this study, we developed AGBE, a novel dual-deaminase-mediated system, that provides a versatile tool for the simultaneous introduction of 4 kinds of base conversions (C-to-G, C-to-T, C-to-A and A-to-G) as well as indels with sgRNAs at the same DNA strand in mammalian immortalized cells, primary somatic cells and embryos.
We constructed 7 versions of AGBE with different components. AGBE-1, containing a rat APOBEC1(R33A) and an ecTadA-TadA*7.10, achieved mainly A-to-G base substitution and almost no conversion of Cs. One reason for this observation was that cytosine deaminase and adenine deaminase could compete for the same target site, resulting in suppression of the editing activity of cytosine deaminase by adenine deaminase (19,21). However, AGBE-2, containing human-derived APOBEC3A instead of rat APOBEC1, produced more abundant gene editing types because, as shown in previous reports, human APOBEC3A-conjugated BEs mediated higher C-to-T base editing than rAPOBEC1 (30–34). Therefore, all six AGBEs (AGBE-2, AGBE-3, AGBE-4, miniAGBE-2, minAGBE-3 and minAGBE-4) with hAPOBEC3A produced simultaneous C-to-G/T/A and A-to-G conversions. The editing efficiencies of AGBE-2 with the ecTadA-TadA*7.10 heterodimer were much lower than those of the original ABEmax, which might be because the function of adenine deaminase was also disturbed by the competition of cytosine deaminase in the AGBE (17). It was reported that the two adenine deaminase mutants ABE8e and ABE8e(V106W) could increase activity (kapp) 590-fold compared with that of ABE7.10 (36,38). Therefore, AGBE-3 and AGBE-4, derived from ecTadA8e and ecTadA8e(V106W), respectively, could result in a higher A-to-G conversion efficiency, comparable to that of ABEmax alone. In the original CGBE1, UNG was used to replace UGI to enhance the C-to-G conversion efficiency (22). However, as reported previously (22,25), we also found that the presence of eUNG in AGBEs had almost no effect on editing frequencies. The amount of endogenous UNG appeared sufficient to meet the demand for excision of the U base. Hence, to reduce the size of AGBE, three smaller AGBEs, namely, miniAGBE-2, miniAGBE-3, and miniAGBE-4 were constructed by removing of the eUNG domain. We expected these miniAGBEs to be delivered into cells more easily than the original AGBEs. In addition, it was reported that ecTadA8e(V106W) could reduce off-target efficiency (36,38). Taking all factors into consideration, miniAGBE-4 without eUNG but containing hAPOBEC3Ai, ecTadA8e(V106W) and nCas9(D10A) was considered the best editing tool to perform saturated mutagenesis.
For A-to-G conversion, the optimal AGBEs not only maintained the same editing efficiency but also maintained editing windows (from positions 4–8) similar to those of ABEs. For C base editing, the editing window expanded from positions 4–8 to 3–13, possibly because the cytosine deaminase (hAPOBEC3A) used in this study could induce a significantly enlarged editing window compared with that of rAPOBEC1 in CGBE1/miniCGBE1 (31,32,34). The expanded editing window was likely an advantage for generating saturated mutagenesis since it increased the abundance of editing outcomes. C-to-G, C-to-T and C-to-A conversions were induced by all versions of AGBEs with hAPOBEC3Ai at all sites, but were induced with different frequencies for a specific mutation at different sites. The frequencies of C-to-G and C-to-T conversions were equal but higher than that of C-to-A conversion. Due to the random editing of Cs and the expanded editing windows within the protospacer, the AGBEs yielded diverse mutation types with different combinations of nucleotide conversions in the same target site guided by a single sgRNA. These included one-base conversions as well as combinations of two to four types of base substitutions, which enriched the gene-editing diversity for the establishment of a saturated mutant population. For example, using hABE site 1 confirmed these capacities of AGBEs because the number of SNV reads induced by all six hAPOBEC3Ai-based AGBEs (AGBE-2–AGBE-4 and miniAGBE-2–miniAGBE-4) was dramatically higher than the number induced by both ABEmax + miniCGBE and ABEmax + CGBE. Similar to the original CGBE system (22,23), the AGBE system generated indels in target sites, but the efficiency of hAPOBEC3Ai-derived AGBEs was substantially higher than that of CGBEs. We hypothesize that when AGBEs function, a nick on one DNA strand is generated by AP lyase or spontaneous lysis of the AP site in the absence of UGI, and a double-strand break (DSB) is then formed with the other nick generated by nCas9(D10A) on the other strand at a nearby location, which results in promotion of the formation of indels through non-homologous end joining (NHEJ). An increased frequency of indels conferred AGBEs with a supplementary capacity to identify target genes related to phenotypes, similar to CRISPR/Cas9.
In addition to determining on-target editing efficiency, evaluating off-target editing risk is an important consideration for the practical application of the AGBE system. To address off-target issues, ABE8e(V106W), an ABE variant with capacity to reduce both RNA and DNA off-target editing (36,38), was used to construct the AGBE with miniCGBE, which itself was confirmed to induce less sgRNA-dependent DNA off-target editing than BE4max (22). All WGS and RNA-seq data showed no significant difference in the number of de novo SNVs and indels among cells expressing miniAGBE-4 with or without sgRNAs and wildtype cells. Therefore, miniAGBE-4 can minimize both DNA and RNA off-target editing activity and can be considered a safe base editor.
Additionally, we found that the AGBEs efficiently generated diverse mutations with the same sgRNA in early-developmental stage embryos, indicating that AGBE could be used to generate animals with saturated mutations in a specific gene when the embryos were transferred into a surrogate. The animals with saturated mutations can be used as animal models containing a substitution mutation-aided lineage-trancing system, which would enrich bio-barcodes with SNVs for mapping the cell phylogeny in vivo. This also meant that AGBEs could be a potential alternative approach for the large-scale production of animals with a variety of mutations for discovering novel phenotypes of mutants in a specific gene, similar to that of N-ethyl-N-nitrosourea (ENU)-induced mutagenesis, which had been used to define novel gene functions at the whole-genome level in Caenorhabditis elegans, flies, zebrafish, mice and even in pigs (50).
To validate the practical applications of AGBEs for the generation of saturated mutant populations, we employed miniAGBE-4 to introduce mass mutations in hDTR and generated corresponding cell clones with different DT sensitivities. As many as 59,269 mutation reads were produced by miniAGBE-4 with 20 sgRNAs targeting the same gene. Among these mutations, we identified some amino acid residues, including Cys121, Tyr123, Glu141 (51) and Thr160, that possibly played key roles in conferring DT insensitivity to human cells. These findings indicate that AGBEs can be used for the directed mutagenesis of drug target genes, which will aid the characterization of drug sensitivity-associated SNVs for the precise treatment of genetic diseases in clinical practice.
In summary, AGBEs can be used to increase the gene-editing diversity for the establishment of saturated mutant populations to verify the functions and consequences of multiple gene mutation patterns through screening.
DATA AVAILABILITY
All the Amplicon deep sequencing and RNA-seq dataset described in this study have been deposited in the GEO database of NCBI (accession number: PRJNA824995). WGS and RNA-seq datasets described in this study have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics (BIG) Data Center (accession number: HRA002278 and HRA002280).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yahai Shu from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Science for providing technical assistance of cell sorting. We are also grateful to the anonymous reviewers of NAR for their extensive and thoughtful suggestions and comments.
Author contributions: L.Lai and K.W. designed and supervised the study. Y.L., J.X., Q.Z. and X.W. performed most of the experiments and data analysis. S.G., L.Lin and T.C. performed the bioinformatics analysis. Q.Z., W.G., F.C., N.L., Z.O. and C.L. performed porcine embryos experiments. Z.Z., M.L., X.L, L.Li, Y.Y. and H.W. provided technical assistance. Y.L., K.W. and L.Lai wrote the manuscript.
Contributor Information
Yanhui Liang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jingke Xie, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Quanjun Zhang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China.
Xiaomin Wang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China.
Shixue Gou, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China.
Lihui Lin, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China.
Tao Chen, Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Weikai Ge, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Zhenpeng Zhuang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Meng Lian, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China.
Fangbing Chen, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Nan Li, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Zhen Ouyang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Chengdan Lai, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Xiaoyi Liu, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lei Li, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yinghua Ye, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China.
Han Wu, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China.
Kepin Wang, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
Liangxue Lai, China-New Zealand Joint Laboratory on Biomedicine and Health, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences (2019RU015), Guangzhou 510530, China; Sanya institute of Swine resource, Hainan Provincial Research Centre of Laboratory Animals, Sanya 572000, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China; Guangdong Provincial Key Laboratory of Large Animal models for Biomedicine, Wuyi University, Jiangmen 529020, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81941004, 32170542]; National Key Research and Development Program of China [2021YFA0805903, 2017YFA0105103, 2018YFB1404205]; Key Research & Development Program of Hainan Province [ZDYF2021SHFZ052]; Major Science and Technology Project of Hainan Province [ZDKJ2021030]; Biological Resources Programme, Chinese Academy of Sciences [KFJ-BRP-017-57]; Youth Innovation Promotion Association of the Chinese Academy of Sciences [2019347]; Key Research & Development Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) [2018GZR110104004]; Science and Technology Planning Project of Guangdong Province, China [2020B1212060052, 2021B1212040016]; Science and Technology Program of Guangzhou, China [202007030003]; 2020 Research Program of Sanya Yazhou Bay Science and Technology City [202002011]; Research Unit of Generation of Large Animal Disease Models, Chinese Academy of Medical Sciences [2019-I2M-5-025]; Young Elite ScientistSponsorship Program by CAST [YESS20200024]. Funding for open access charge: Science and Technology Program of Guangzhou, China (202007030003).
Conflict of interest statement. None declared.
REFERENCES
- 1. Chaudhary R., Singh B., Kumar M., Gakhar S.K., Saini A.K., Parmar V.S., Chhillar A.K.. Role of single nucleotide polymorphisms in pharmacogenomics and their association with human diseases. Drug Metab. Rev. 2015; 47:281–290. [DOI] [PubMed] [Google Scholar]
- 2. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J.et al.. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun Y.V., Hu Y.J.. Integrative analysis of Multi-omics data for discovery and functional studies of complex human diseases. Adv. Genet. 2016; 93:147–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morgil H., Can Gercek Y., Tulum I.. Single nucleotide polymorphisms (SNPs) in plant genetics and breeding: Genetic Polymorphisms. 2020; 1–12. [Google Scholar]
- 5. Rothschild. M.F., Hu. Z.-l., Jiang. Z. Advances in QTL mapping in pigs. Int. J. Biol. Sci. 2007; 3:192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma L., Boucher J.I., Paulsen J., Matuszewski S., Eide C.A., Ou J., Eickelberg G., Press R.D., Zhu L.J., Druker B.J.et al.. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canver M.C., Tripathi P., Bullen M.J., Olshansky M., Kumar Y., Wong L.H., Turner S.J., Lessard S., Pinello L., Orkin S.H.et al.. A saturating mutagenesis CRISPR-Cas9-mediated functional genomic screen identifies cis- and trans-regulatory elements of oct4 in murine ESCs. J. Biol. Chem. 2020; 295:15797–15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W.. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014; 509:487–491. [DOI] [PubMed] [Google Scholar]
- 9. Shalem O., Sanjana N.E., Zhang F.. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015; 16:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weber J., Ollinger R., Friedrich M., Ehmer U., Barenboim M., Steiger K., Heid I., Mueller S., Maresch R., Engleitner T.et al.. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13982–13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuella-Martin R., Hayward S.B., Fan X., Chen X., Huang J.W., Taglialatela A., Leuzzi G., Zhao J., Rabadan R., Lu C.et al.. Functional interrogation of DNA damage response variants with base editing screens. Cell. 2021; 184:1081–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanna R.E., Hegde M., Fagre C.R., DeWeirdt P.C., Sangree A.K., Szegletes Z., Griffith A., Feeley M.N., Sanson K.R., Baidi Y.et al.. Massively parallel assessment of human variants with base editor screens. Cell. 2021; 184:1064–1080. [DOI] [PubMed] [Google Scholar]
- 13. Xu P., Liu Z., Liu Y., Ma H., Xu Y., Bao Y., Zhu S., Cao Z., Wu Z., Zhou Z.et al.. Genome-wide interrogation of gene functions through base editor screens empowered by barcoded sgRNAs. Nat. Biotechnol. 2021; 39:1403–1413. [DOI] [PubMed] [Google Scholar]
- 14. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y.et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016; 353:aaf8729. [DOI] [PubMed] [Google Scholar]
- 16. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li C., Zhang R., Meng X., Chen S., Zong Y., Lu C., Qiu J.L., Chen Y.H., Li J., Gao C.. 2020) Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38:875–882. [DOI] [PubMed] [Google Scholar]
- 18. Grunewald J., Zhou R., Lareau C.A., Garcia S.P., Iyer S., Miller B.R., Langner L.M., Hsu J.Y., Aryee M.J., Joung J.K.. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020; 38:861–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakata R.C., Ishiguro S., Mori H., Tanaka M., Tatsuno K., Ueda H., Yamamoto S., Seki M., Masuyama N., Nishida K.et al.. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020; 38:865–869. [DOI] [PubMed] [Google Scholar]
- 20. Zhang X., Zhu B., Chen L., Xie L., Yu W., Wang Y., Li L., Yin S., Yang L., Hu H.et al.. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020; 38:856–860. [DOI] [PubMed] [Google Scholar]
- 21. Xie J., Huang X., Wang X., Gou S., Liang Y., Chen F., Li N., Ouyang Z., Zhang Q., Ge W.et al.. ACBE, a new base editor for simultaneous C-to-T and A-to-G substitutions in mammalian systems. BMC Biol. 2020; 18:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurt I.C., Zhou R., Iyer S., Garcia S.P., Miller B.R., Langner L.M., Grunewald J., Joung J.K.. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2020; 39:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao D., Li J., Li S., Xin X., Hu M., Price M.A., Rosser S.J., Bi C., Zhang X.. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2020; 39:35–40. [DOI] [PubMed] [Google Scholar]
- 24. Chen L., Park J.E., Paa P., Rajakumar P.D., Prekop H.T., Chew Y.T., Manivannan S.N., Chew W.L.. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021; 12:1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koblan L.W., Arbab M., Shen M.W., Hussmann J.A., Anzalone A.V., Doman J.L., Newby G.A., Yang D., Mok B., Replogle J.M.et al.. Efficient C*G-to-G*C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 2021; 39:1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindahl T. An N-glycosidase from escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai L., Kolber-Simonds D., Park K.W., Cheong H.T., Greenstein J.L., Im G.S., Samuel M., Bonk A., Rieke A., Day B.N.et al.. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002; 295:1089–1092. [DOI] [PubMed] [Google Scholar]
- 28. Yang D., Wang C.E., Zhao B., Li W., Ouyang Z., Liu Z., Yang H., Fan P., O’Neill A., Gu W.et al.. Expression of huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum. Mol. Genet. 2010; 19:3983–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S.. EditR: a method to quantify base editing from sanger sequencing. CRISPR J. 2018; 1:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X., Li J., Wang Y., Yang B., Wei J., Wu J., Wang R., Huang X., Chen J., Yang L.. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 2018; 36:946–949. [DOI] [PubMed] [Google Scholar]
- 31. Zong Y., Song Q., Li C., Jin S., Zhang D., Wang Y., Qiu J.L., Gao C.. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018; 36:950–953. [DOI] [PubMed] [Google Scholar]
- 32. Liu Z., Chen S., Shan H., Zhang Q., Chen M., Lai L., Li Z.. Efficient and precise base editing in rabbits using human APOBEC3A-nCas9 fusions. Cell Discov. 2019; 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X., Ding C., Yu W., Wang Y., He S., Yang B., Xiong Y.C., Wei J., Li J., Liang J.et al.. Cas12a base editors induce efficient and specific editing with low DNA damage response. Cell Rep. 2020; 31:107723. [DOI] [PubMed] [Google Scholar]
- 34. Lian M., Chen F., Huang X., Zhao X., Gou S., Li N., Jin Q., Shi H., Liang Y., Xie J.et al.. Improving the Cpf1-mediated base editing system by combining dCas9/dead sgRNA with human APOBEC3A variants. J. Genet. Genomics. 2021; 48:92–95. [DOI] [PubMed] [Google Scholar]
- 35. Ding D., Chen K., Chen Y., Li H., Xie K.. Engineering introns to express RNA guides for Cas9- and Cpf1-mediated multiplex genome editing. Mol. Plant. 2018; 11:542–552. [DOI] [PubMed] [Google Scholar]
- 36. Rees H.A., Wilson C., Doman J.L., Liu D.R.. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 2019; 5:eaax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lapinaite A., Knott G.J., Palumbo C.M., Lin-Shiao E., Richter M.F., Zhao K.T., Beal P.A., Liu D.R., Doudna J.A.. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science. 2020; 369:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E.et al.. Phage-assisted evolution of an adenine base editor with improved cas domain compatibility and activity. Nat. Biotechnol. 2020; 38:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grunewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K.. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019; 569:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R.et al.. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019; 571:275–278. [DOI] [PubMed] [Google Scholar]
- 42. Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H.. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019; 364:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L, Zhang F.et al.. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019; 364:292–295. [DOI] [PubMed] [Google Scholar]
- 44. Bae S., Park J., Kim J.S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pappenheimer A.M. Jr, Harper A.A., Moynihan M., Brockes J.P. Diphtheria toxin and related proteins: effect of route of injection on toxicity and the determination of cytotoxicity for various cultured cells. J. Infect. Dis. 1982; 145:94–102. [DOI] [PubMed] [Google Scholar]
- 46. Mitamura. T., Higashiyama. S., Taniguchi. N., Klagsbrun. M., Mekada. E. Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGF-like growth factor diphtheria toxin receptor and inhibits specifically its mitogenic activity. J. Biol. Chem. 1995; 270:1015–1019. [DOI] [PubMed] [Google Scholar]
- 47. Wang M., Marin A.. Characterization and prediction of alternative splice sites. Gene. 2006; 366:219–227. [DOI] [PubMed] [Google Scholar]
- 48. Xue C., Zhang H., Lin Q., Fan R., Gao C.. Manipulating mRNA splicing by base editing in plants. Sci. China Life Sci. 2018; 61:1293–1300. [DOI] [PubMed] [Google Scholar]
- 49. Yuan J., Ma Y., Huang T., Chen Y., Peng Y., Li B., Li J., Zhang Y., Song B., Sun X.et al.. Genetic modulation of RNA splicing with a CRISPR-Guided cytidine deaminase. Mol. Cell. 2018; 72:380–394. [DOI] [PubMed] [Google Scholar]
- 50. Hai T., Cao C., Shang H., Guo W., Mu Y., Yang S., Zhang Y., Zheng Q., Zhang T., Wang X.et al.. Pilot study of large-scale production of mutant pigs by ENU mutagenesis. Elife. 2017; 6:e26248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mitamura T., Umata T., Nakano F., Shishido Y., Toyoda T., Itai A., Kimura H., Mekada E.. Structure–function analysis of the diphtheria toxin receptor toxin binding site by site-directed mutagenesis. J. Biol. Chem. 1997; 272:27084–27090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the Amplicon deep sequencing and RNA-seq dataset described in this study have been deposited in the GEO database of NCBI (accession number: PRJNA824995). WGS and RNA-seq datasets described in this study have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics (BIG) Data Center (accession number: HRA002278 and HRA002280).