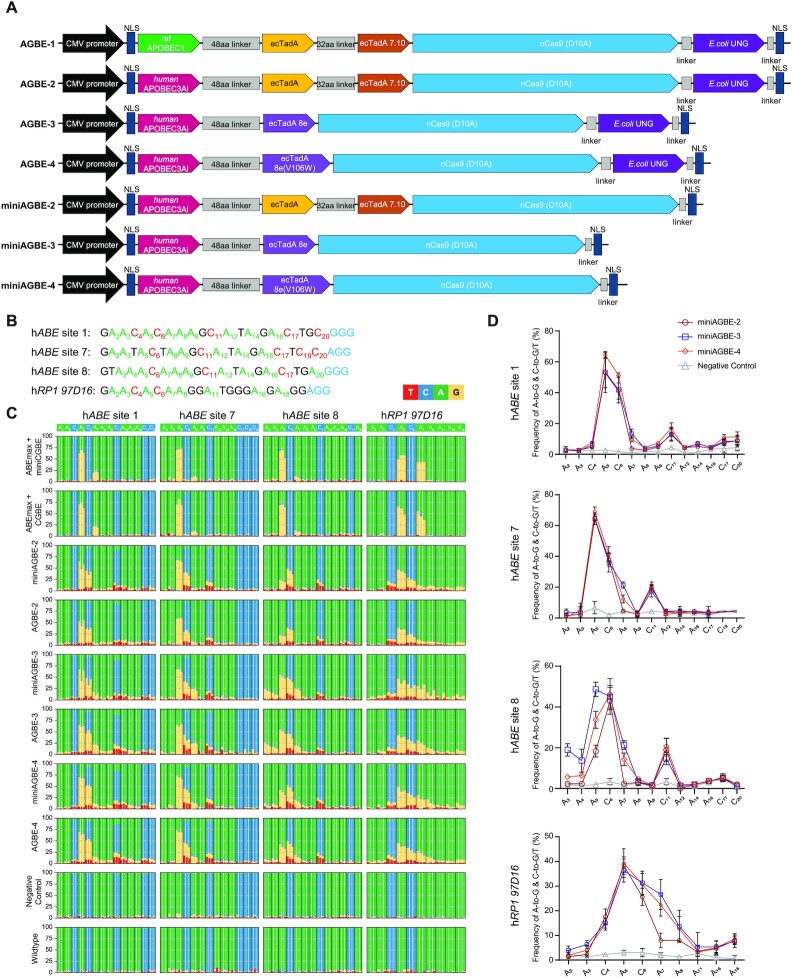
Figure 2.
Base editing activities of AGBEs with different architectures in HEK293 cells. (A) Architectures of miniAGBEs and AGBEs. aa, amino acid; NLS, nuclear localization signal. (B) Protospacers and PAM (blue) sequences of four human genomic loci studied by AGBEs, with target As (green) and Cs (red) in (C). (C) Bar plots shows on-target As (green) and Cs (blue) base editing frequencies induced by various AGBEs with four sgRNAs targeting genomic loci in HEK293 cells. Editing frequencies of three independent replicates at each base are display side-by-side. (D) Comparison of A-to-G and C-to-G/C-to-T editing frequencies induced by three miniAGBEs (The indistinctive conversion of C-to-A are not shown). Values and error bars indicate the mean ± s.d. of three independent replicates. Subscript number indicates position of the base in the protospacer, counting the PAM as position 21–23. HEK293 cells electro-transfected with sgRNA only served as negative control group, and cells electro-transfected with resuspension buffer only served as WT group. The editing efficiency in (C and D) is analysed by EditR for quantification.