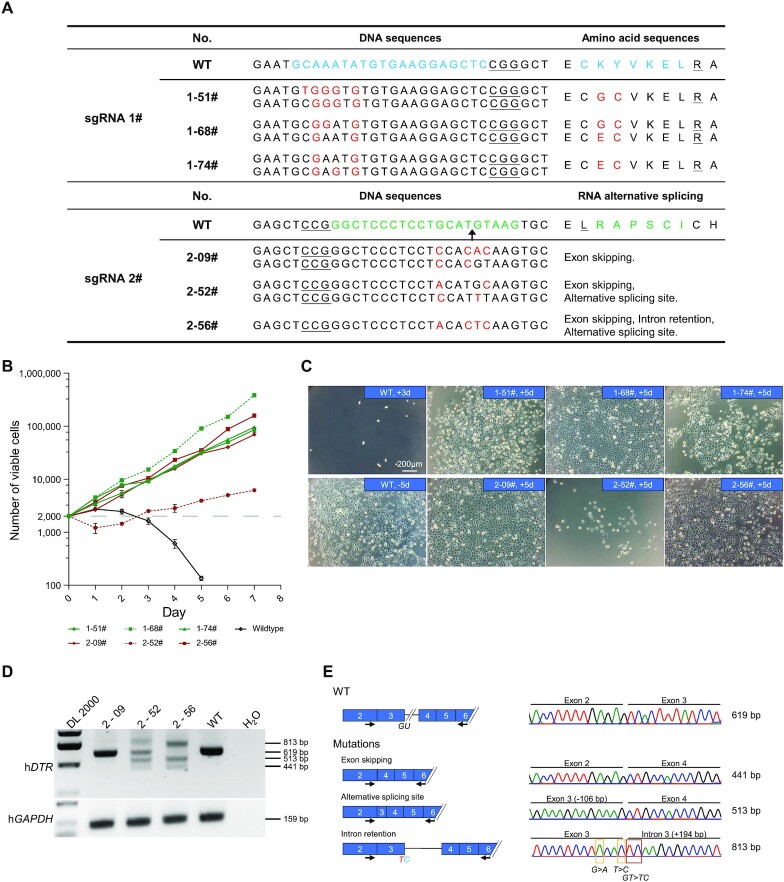
Figure 6.
Characterization of the function of hDTR variants generated by miniAGBE-4. (A) Genotypes and corresponding amino acid substitution (sgRNA-1) or RNA alternative splicing events (sgRNA-2) of hDTR-mutated single-cell clones derived from miniAGBE-4/sgRNA-1 and miniAGBE-4/sgRNA-2 edited HEK293 cells. Target DNA sequence and amino acids (blue and green), PAM (underline), mutant sites and amino acids (red). (B) Cell proliferation in DT-supplemented medium. Wildtype HEK293 cells are almost completely eliminated by DT after 3 days culturing. Values and error bars indicate the mean ± s.d. of three independent replicates. (C) Images of DT-insensitive single-cell clones with DT treatment for 5 days. +3d/+5d, cultured in DT-supplemented medium for 3 or 5 days. -5d, cultured in normal medium for 5 days. Scale bar: 200 μm. (D) RT-PCR analysis of isoforms expression in WT HEK293 cells and single-cell clones derived from miniAGBE-4/sgRNA-2-edited cells. Data are the representative of three independent replicates. DL 2000, DNA marker. (E) Schematic diagram of hDTR mRNA variants in WT and mutated cell clones edited the 5′ splice site of exon 3 in hDTR and details of the sequences of RT-PCR amplicons.