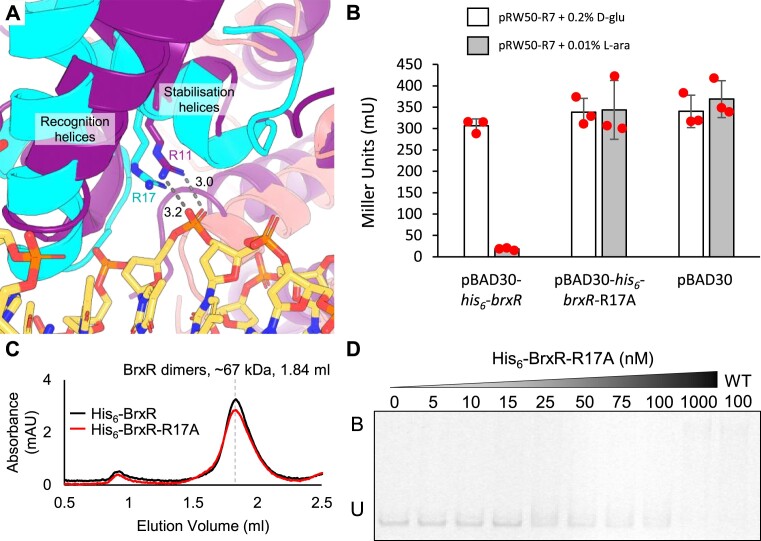
Figure 5.
BrxREfer R17 is essential for transcriptional repression and DNA binding. (A) Close-up alignment of wHTH domains from the BrxREfer apo structure (cyan and salmon cartoons, PDB: 7QFZ), and BrxRAcin-DNA structure (purple cartoon and DNA as sticks, PDB: 7T8K). Stabilization and recognition helices roughly overlay, and there is co-localization of BrxREfer R17 with BrxRAcin R11. BrxRAcin R11 makes bidentate hydrogen bonds with the DNA phosphate backbone. Distances shown are in angstroms. (B) LacZ-reporter assays using active pRW50-R7 construct with and without induction of His6-BrxREfer or His6-BrxREfer-R17A from pBAD30. Data are shown in triplicate, and error bars represent standard deviation of the mean. (C) Size exclusion chromatography of His6-BrxREfer and His6-BrxREfer-R17A resolved via a Superdex 200 increase GL 5/150 gel filtration column. Both proteins eluted at 1.84 ml, corresponding to a mass approximately twice their respective Mr, indicating correct folding into dimer formation. No additional peak was observed for residual monomers. (D) Electrophoretic mobility shift assay (EMSA) of titrated His6-BrxREfer-R17A protein with WT dsDNA probe IR1-IR2 spanning pEFER nucleotide locations 12,801–12,870. Target probe was amplified to incorporate fluorescein and contains the native promoter region. Protein concentration is shown above each lane together with binding events (B – bound, U – unbound). Control lane of His6-BrxR (WT) is included for comparison. Experiment was run in triplicate and a representative gel is shown.