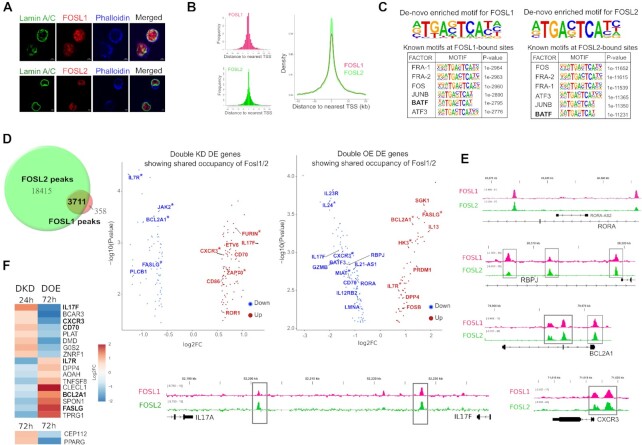
Figure 4.
Genome-wide occupancy profile of FOSL proteins in human Th17 cells. (A) Immunofluorescence images showing nuclear localization of FOSL1 (red, above panel) and FOSL2 (red, below panel) in 72 h-polarized Th17 cells. Lamin A/C (in green) marks the nuclear periphery, whereas phalloidin (in blue) stains the cytoplasmic actin. (B) ChIP-seq analysis was performed for FOSL1 and FOSL2 using 72 h-cultured Th17 cells. Figures on the left show distribution of FOSL1 and FOSL2 binding sites relative to the position of the closest transcription start site (TSS). TSS is defined to be at position zero. Figure on the right shows an overlay of the peak distribution profiles of the two factors. (C) The topmost consensus sequences for FOSL1 and FOSL2 genomic binding were identified using de-novo motif enrichment analysis by Homer. FOSL1 (left) and FOSL2 (right) peaks were further enriched for known TF motifs, and the top motifs identified by Homer are shown. Peaks with IDR p < 0.01 were used for motif discovery. (D) ChIPpeakAnno was used to determine the overlap in the genomic binding sites of FOSL1 and FOSL2 (overlap represents peaks sharing 200 bp or more). Genes neighbouring to these overlying sites and differentially expressed under DKD or DOE conditions (FDR ≤ 0.1, |FC| ≥ 1.5) were assigned as the shared-direct targets of FOSL1 and FOSL2. Adjoining volcano plots show the logarithmic fold changes for selected shared targets (DKD (left); DOE (right)). Downregulated genes are in blue, and upregulated ones are in red. Targets with FOSL occupancy over promoter regions (5-kb window around TSS) are marked with asterisk. (E) Integrative Genomics Viewer (IGV) track snapshots show the binding overlap of FOSL1 and FOSL2 over selected Th17-associated genes. (F) Heatmap depicts the shared direct targets that show opposite expression changes in FOSL DKD and DOE conditions, at the indicated time points of Th17 differentiation. Th17-relevant genes have been highlighted.