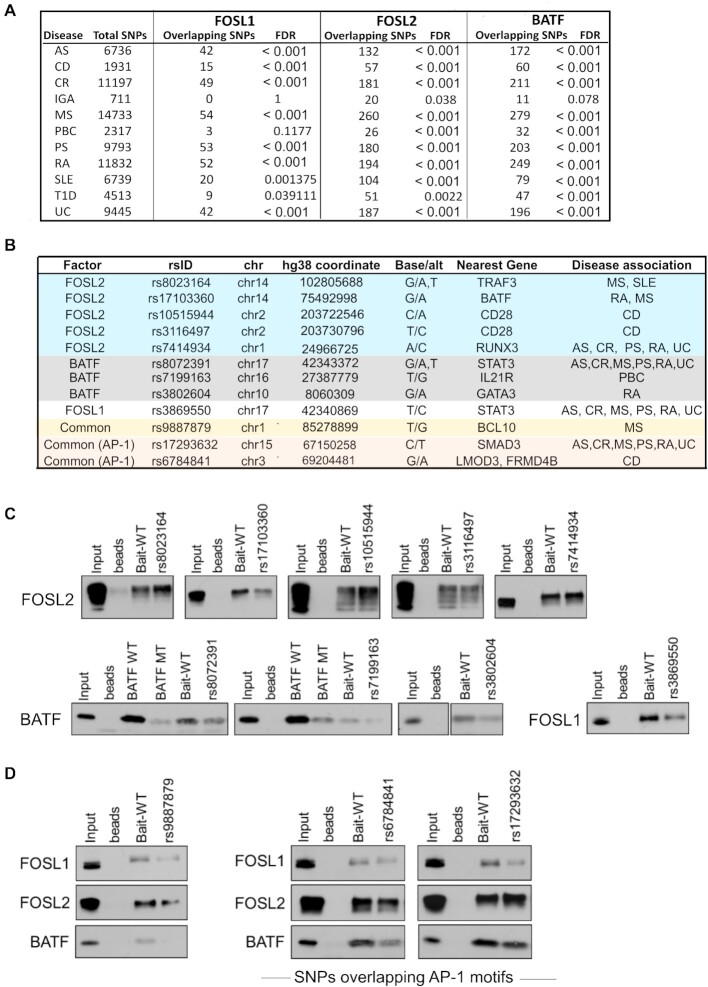
Figure 8.
SNPs associated with autoimmune diseases localize within the genomic binding sites of FOSL1, FOSL2 and BATF. (A) Enrichment of disease-associated SNPs (or their proxies in Caucasian populations) within FOSL1, FOSL2 and BATF genomic-binding sites, relative to random sets of background SNPs. (B) SNPs relevant to the study were shortlisted (Supplementary Table S8). Of these, the SNPs that were functionally validated in DNA-affinity precipitation assays are shown. (C, D) DAPA followed by immunoblot analysis shows the SNPs that alter the binding of FOSL1, FOSL2 or BATF to their genomic sites (identified by ChIP-seq analysis). Wildtype (WT) oligonucleotides containing the binding motifs of these TFs (at different genomic loci), and mutant oligonucleotides harbouring a SNP within the binding motif, were used as baits for pull-down of the corresponding AP-1 factor from 72 h Th17-polarized cell lysates. For experimental controls, an oligonucleotide with a conserved binding sequence for BATF (BATF WT), and the corresponding mutated sequence which is known to disrupt BATF occupancy (BATF MUT) were used. Panel C includes SNPs affecting the binding of either FOSL1, FOSL2 or BATF. Those SNPs at the common binding sites of the three factors which also alter the binding affinities for all of them are shown in panel D. The common SNPs harboured within consensus AP-1 motifs are labelled. Data is representative of three biological replicates.