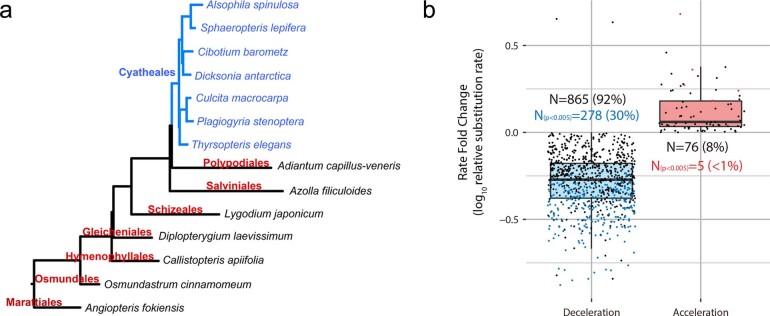
Extended Data Fig. 4. Genome-wide deceleration of nucleotide substitution rate in Cyatheales ferns.
(a) Phylogenetic tree generated by OrthoFinder depicts the relationships of 14 species from eight orders, including Cyatheales, Polypodiales, Salviniales, Schizeales, Gleicheniales, Hymenophyllales, Osmundales and Marattiales. The branch lengths within Cyatheales are shorter than those in other orders, suggesting deceleration of substitution rate in Cyatheales. (b) Genome-wide substitution rate variation in Cyatheales (N is the number of nuclear protein-coding genes). Among the 941 single-copy nuclear genes from Cyatheales, a majority (92%) showed reduced substitution rates, and the reduction in 30% genes was statistically significant (p <0.005). By contrast, <1% of genes had significant elevated rates. Upper bound of each box (Q3) represents the 75th percentile, lower bound of each box (Q1) represents the 25th percentile, the midline of each box is the median, and each whisker represents the highest or lowest point within Q3 + 1.5*IQR or Q1 - 1.5*IQR, respectively (IQR = Q3 - Q1). P-values were calculated using an one-sided likelihood-ratio test.