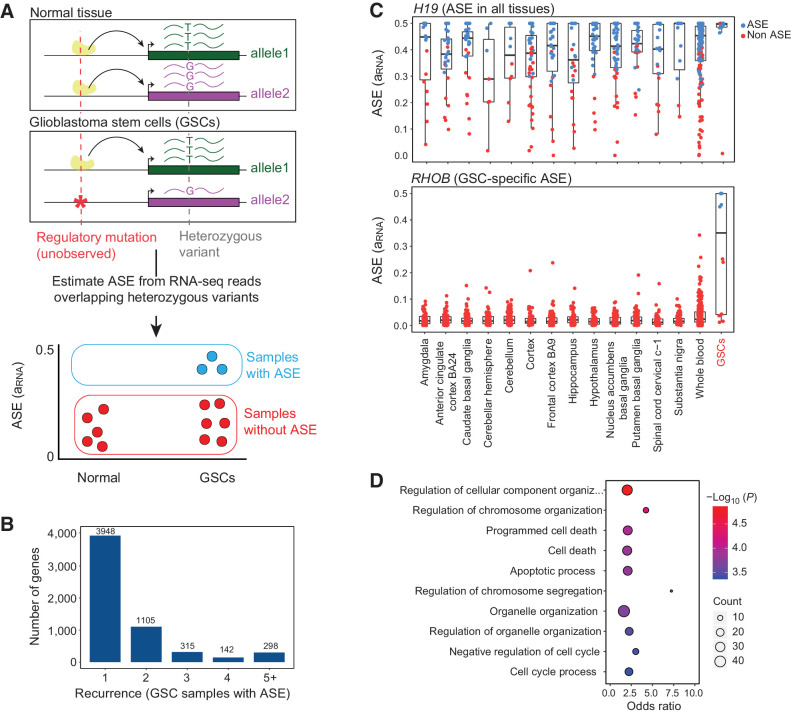
Figure 1.
Discovery of genes with recurrent allele-specific expression in GSCs. A, Schematic of approach. ASE is the higher expression of one allele of a gene compared with the other allele and can be used to detect the effects of cis-regulatory mutations, even when the cis-regulatory mutation is unobserved. ASE is estimated from RNA-seq reads that overlap heterozygous variants within exons. We identify genes that exhibit ASE in GSCs more frequently than in normal tissues. B, Recurrence of ASE in GSCs. The histogram indicates the number of GSCs with ASE (FDR corrected P ≤ 10%) across 42 patient-derived GSCs, for genes that have ASE in at least one sample. C, Estimated ASE (aRNA) for GSCs and normal brain and whole-blood samples from GTEx for a known imprinted gene, H19 (top) and RHOB (bottom). Each point is a sample. Points are colored based on the significance of ASE (likelihood ratio test FDR ≤ 10%). D, GO analysis of 118 genes that are enriched for ASE in GSCs compared with normal samples. The figure shows the top 10 GO categories.