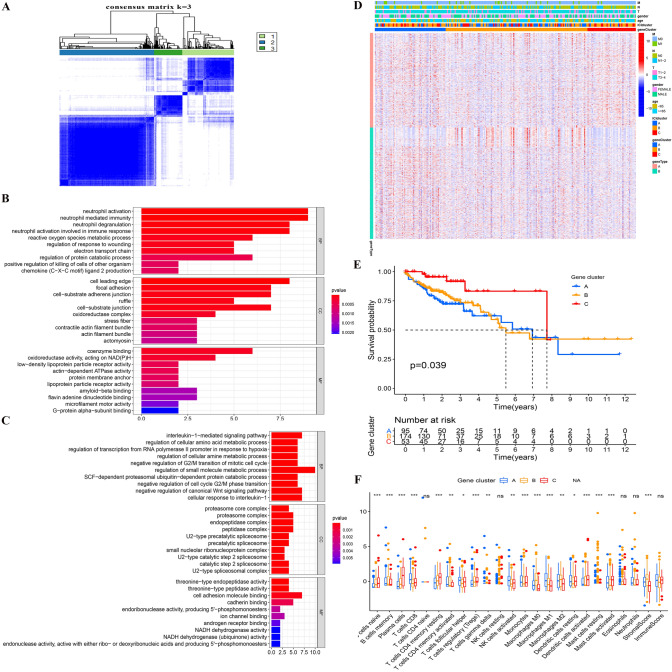
Fig. 2.
| A Consensus matrixes of TCGA-COAD cohorts for appropriate k value (k = 3), displaying the clustering stability using 1000 iterations of hierarchical clustering. TCGA samples were clustered into 3 subtypes based on the DEGs among three ICI clusters. B GO enrichment analysis of the ICI-relevant signature genes A. C GO enrichment analysis of the ICI-relevant signature genes B. The X axis indicated the number of genes within each GO term. D The heat map depicted the expression of DEGs in different ICI clusters and gene clusters. Heat map colors indicate relative DEGs expression levels. E Kaplan–Meier curves of overall survival in different gene clusters. The log rank test showed an overall p = 0.039. F The fraction of tumor-infiltrating immune cells, immune score and stromal score in three gene clusters. The statistical difference of three gene clusters was compared by the Kruskal–Wallis test (*p < 0.05, **p < 0.01, ***p < 0.001, nsp > 0.05)