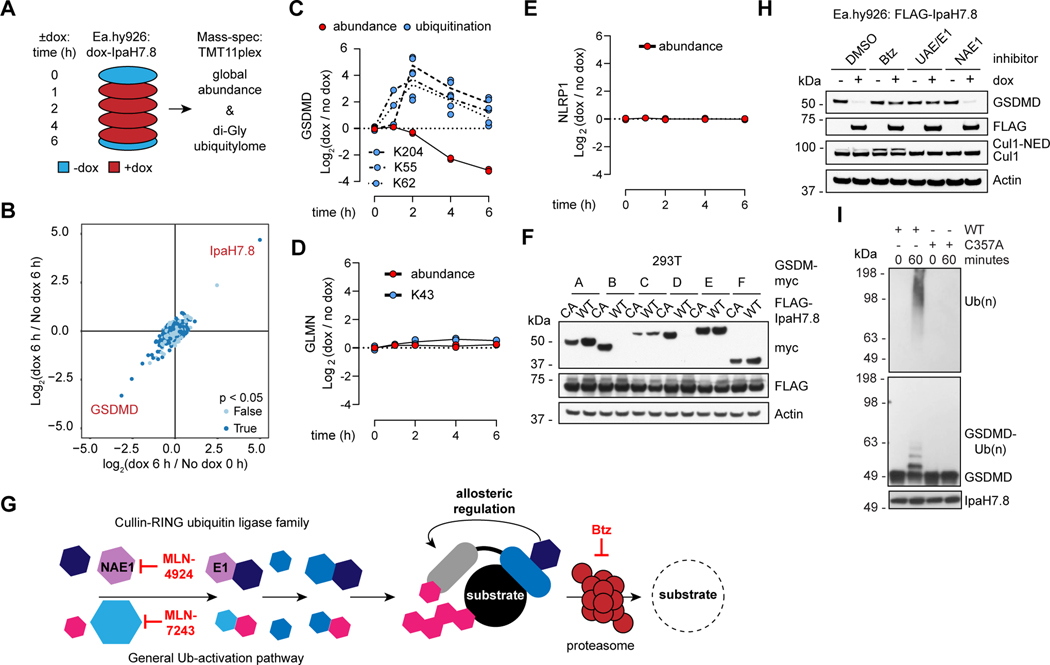
Figure 2. IpaH7.8 targets GSDMD for proteasomal degradation.
(A) Proteomics strategy to identify IpaH7.8 substrate(s) in Ea.hy926 cells.
(B) Scatter plot depicting doxycycline (dox)-induced IpaH7.8-dependent changes in global protein abundance. Results represent 2 biological replicates. The x-axis corresponds to the Log2 ratio of 6 h dox / 0 h no dox. The Log2 ratio of 6 h dox / 6 h no dox is on the y-axis. P-values determined by ANOVA.
(C, D and E) GSDMD (C), GLMN (D), and NLRP1, (E) (red circles) and associated ubiquitinated lysines (blue circles) are quantified at different times in Ea.hy926 cells after doxycycline (dox)-induced expression of IpaH7.8. Each circle represents the Log2 ratio of dox treated / no dox (2 biological replicates each).
(F) Immunoblots of 293T cells co-transfected with myc-tagged gasdermins and Flag-IpaH7.8 (WT, wild-type or CA, mutant C357A). Results representative of 3 independent experiments.
(G) The mammalian ubiquitin pathway. Specific inhibitors of UAE/E1 (MLN-7243), NAE1 (MLN-4924), and the proteasome (Btz) are annotated at the relevant steps. Human cullin-RING family E3 ligases require the co-factor NEDD8 (NED) for activity.
(H) Immunoblots of Ea.hy926 cells with dox-inducible Flag-IpaH7.8 after treatment for 4 h with DMSO vehicle or 1 μM Btz, MLN-7243 (UAE/E1), or MLN-4924 (NAE1).
(I) Immunoblots of in vitro ubiquitination reactions using GSDMD and IpaH7.8 (WT or C357A). Results representative of 3 independent experiments.