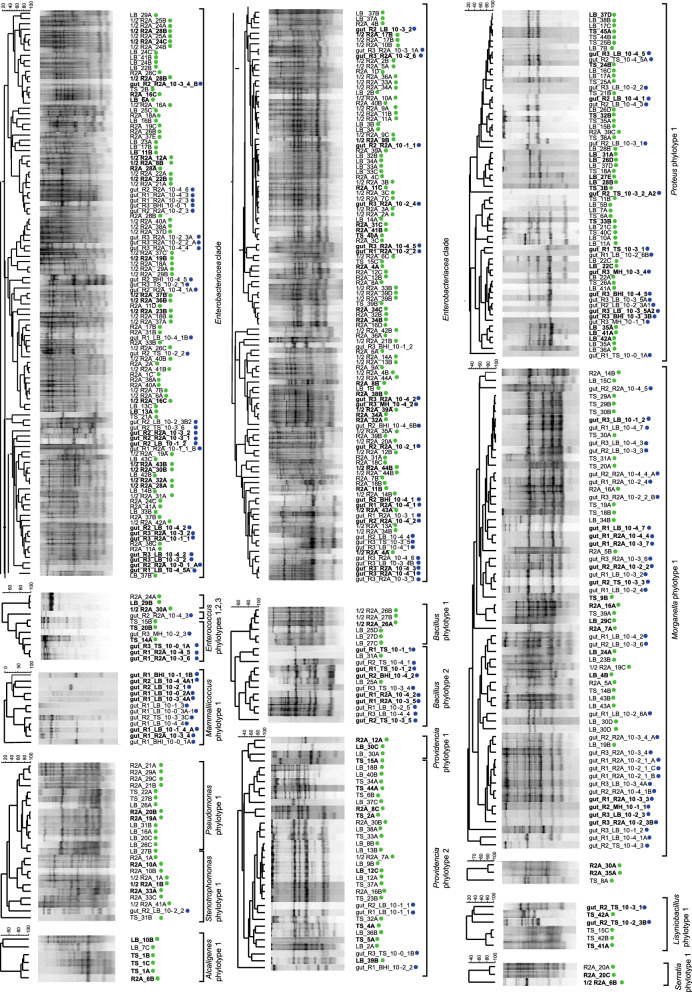
Fig. 4.
Genotyping of all identified phylotypes isolated from BSFL gut microbiota, based on fingerprint pattern obtained by genomic fingerprinting using primers targeting BOX and (GTG)5 repetitive elements. (GTG)5-PCR was employed exclusively with Mammaliicoccus and Enterococcus phylotypes. Cluster analysis was performed in BioNumerics (Applied Maths) using UPGMA clustering, based on a dissimilarity matrices generated by the Pearson correlation. Bold isolates were identified through 16S rRNA gene sequencing. Blue and green dots represent the isolates cultured by direct plating and dilution-to-extinction cultivation. (Color figure online)