FIGURE 1.
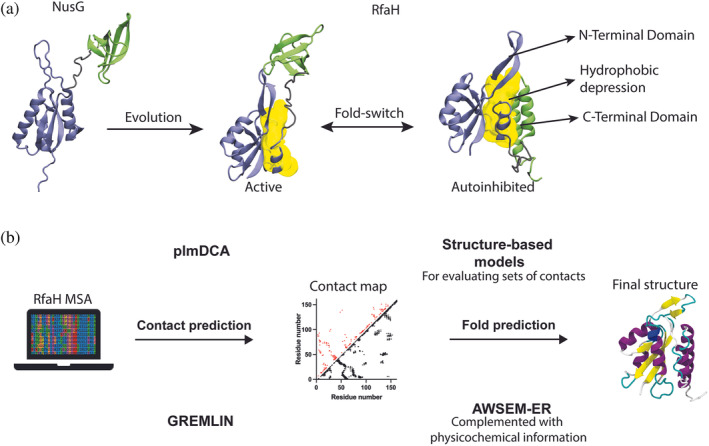
Summary of the research and methods used in this work. (a) NusG, a non‐metamorphic protein, evolved into its paralog RfaH, whose fold‐switch is characterized to take place between an autoinhibited fold (αRfaH) that has interdomain contacts at the hydrophobic patch (yellow) and an active fold that does not establish interdomain contacts (βRfaH). We hypothesized that the emergence of these intradomain and interdomain contacts can be inferred via coevolutionary analysis. (b) By constructing a metagenomic‐enriched multiple sequence alignment (MSA) of RfaH and filtering out non‐metamorphic sequences based on secondary structure predictions, we inferred a contact map of coevolving residue pairs that we used to predict the structures of the autoinhibited and active states of RfaH through molecular dynamics using two different pipelines, namely DCA/SBM and GREMLIN/AWSEM‐ER, which capture the distinctive features of RfaH folding.
