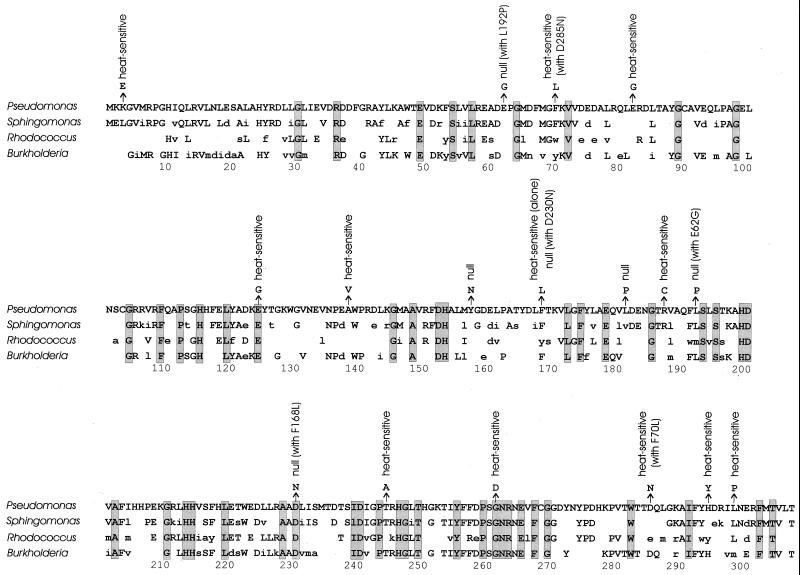
FIG. 4.
Comparison of amino acid substitutions causing defects in Pseudomonas XylE with amino acids conserved during divergence of XylE in Sphingomonas (24), Rhodococcus (2), and Burkholderia (16). This presentation does not take into account gaps required for optimal alignment. Amino acid residues identical to those in Pseudomonas XylE are in uppercase, similar amino acids are in lowercase, and different amino acids are not shown. Shaded boxes mark amino acid residues conserved in all four proteins.