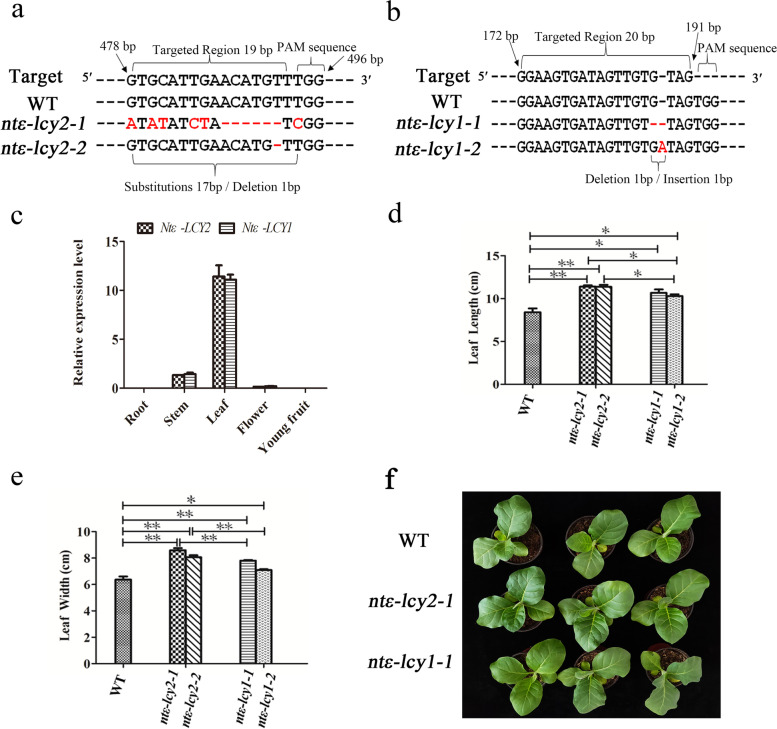
Fig. 1.
Identification and phenotypic analysis of homozygous Ntε-LCY mutants. a–b Sequence analysis of Ntε-LCY2 and Ntε-LCY1 homozygous mutant strains generated by CRISPR/Cas9; the red font indicates the gene editing area. c Tissue expression patterns of Ntε-LCY2 and Ntε-LCY1 in roots, stems, leaves, flowers and young fruit by qRT-PCR. Data are presented as the mean ± standard error of the mean (SEM) of three independent experiments (n = 3; two-way ANOVA; Tukey’s post-hoc test). d–e Maximum length and width of the third true leaf (L3) of five tobacco materials (WT, ntε-lcy2-1, ntε-lcy2-2, ntε-lcy1-1, and ntε-lcy1-2). Data are presented as the mean ± SEM of at least 12 plants. Scale bar = 1 cm (n = 12 *p < 0.05, **p < 0.01; one-way ANOVA; Tukey’s post-hoc test). f Phenotypes of three lines (WT, ntε-lcy2-1, and nt ε-lcy1-1), and the interval of each two plants is 15 cm