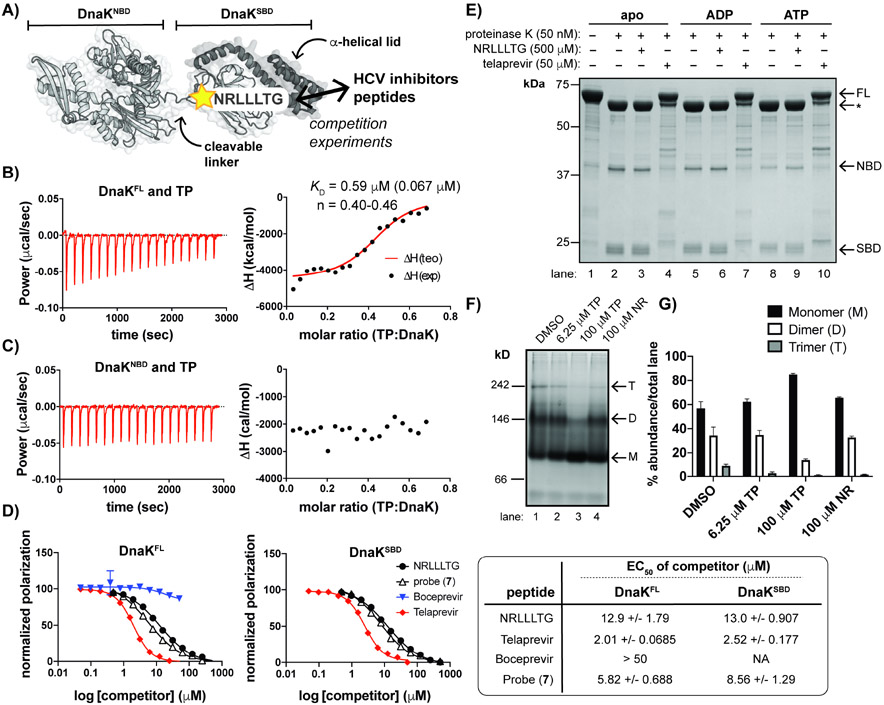
Figure 4. TP interacts with the SBD of DnaK and promotes a monomeric state.
(A) Schematic of DnaK with indicated binding site of NR peptide, a substrate mimic. (B) ITC measurements depict interaction of full-length DnaK (DnaKFL) with TP. KD represents the average of n=3 experiments with SD shown in parentheses; n: stoichiometry value; exp: experimental; teo: theoretical. (C) An N-terminal truncation (DnaKNBD) does not interact with TP under similar conditions to those used in B. (D) Fluorescence polarization (FP) assays demonstrate that TP and an analog (7) displace FL-NR from both DnaKFL (left) and an SBD truncation (DnaKSBD) (right). BC does not compete with FL-NR. 95% confidence intervals of the EC50 values are indicated (far right). (E) Limited proteolysis of apo DnaK +/− excess nucleotides show that samples with TP retained the greatest amount of full-length (FL) DnaK compared to the apo and NR samples. *Indicates partial cleavage of the C-terminal α-helical lid (~60 kDa fragment). (F) Native gel electrophoresis analysis and (G) quantification of replicate experiments (n=3, error bars represent SD) of apo DnaK incubated with a range of [TP] and excess NR indicates that TP shifts DnaK to a monomeric species while NR does not. Oligomeric states are approximated, where M = monomer; D = dimer and T = trimer. Gels and traces are all representative of n = 3 experiments. See also Figures S3 and S4.