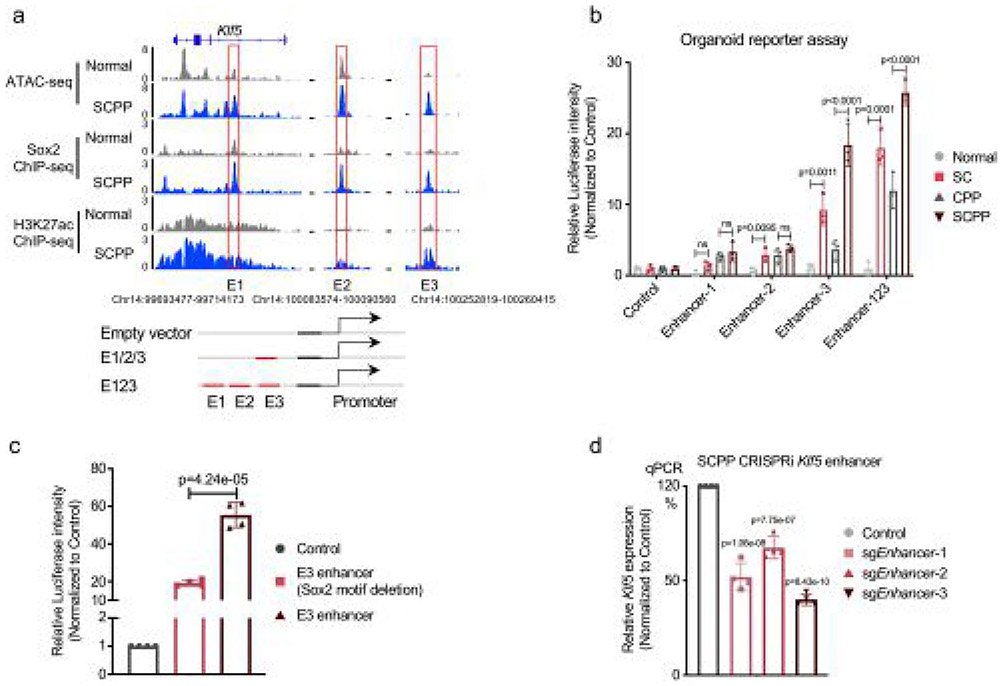
Extended Data Fig. 5 ∣. Sox2 overexpression activates Klf5 by binding to Klf5 enhancer.
a, Sox2 and H3K27ac ChIP-seq and ATAC-seq profiles in Normal and SCPP organoids identify constituent enhancers e1-e3. b, Luciferase reporter assays measuring the enhancer activity of e1-e3 in four groups of organoids. The pGL3 plasmid without the enhancer region (empty) is used as a negative control. Along the Y-axis, relative Luciferase units are normalized to negative control. Three biological replicates were performed for each organoid. Data are shown as Mean ± SD, p-value calculated by one-way ANOVA followed by Benjamini-Hochberg correction. c, Luciferase reporter assays measuring the enhancer activity of E3 with and without the Sox2 motif deleted in SCPP organoids. The pGL3 plasmid without the enhancer region (empty) is used as a negative control. Along the Y-axis, relative Luciferase units are normalized to negative control. Four biological replicates were performed for each organoid. Data are shown as Mean ± SD, p-value calculated by two-sided t-tests. d, mRNA expression of Klf5 in SCPP organoids with and without dCAS9-mediated e1-e3 enhancer repression as measured by RT-PCR. The sgNT is used as a negative control. Four biological replicates were performed. Data are shown as Mean ± SD, p-value calculated by one-way ANOVA followed by Benjamini-Hochberg correction.