Fig. 1. Rationale for a multiscale map of cancer systems and overview of its assembly.
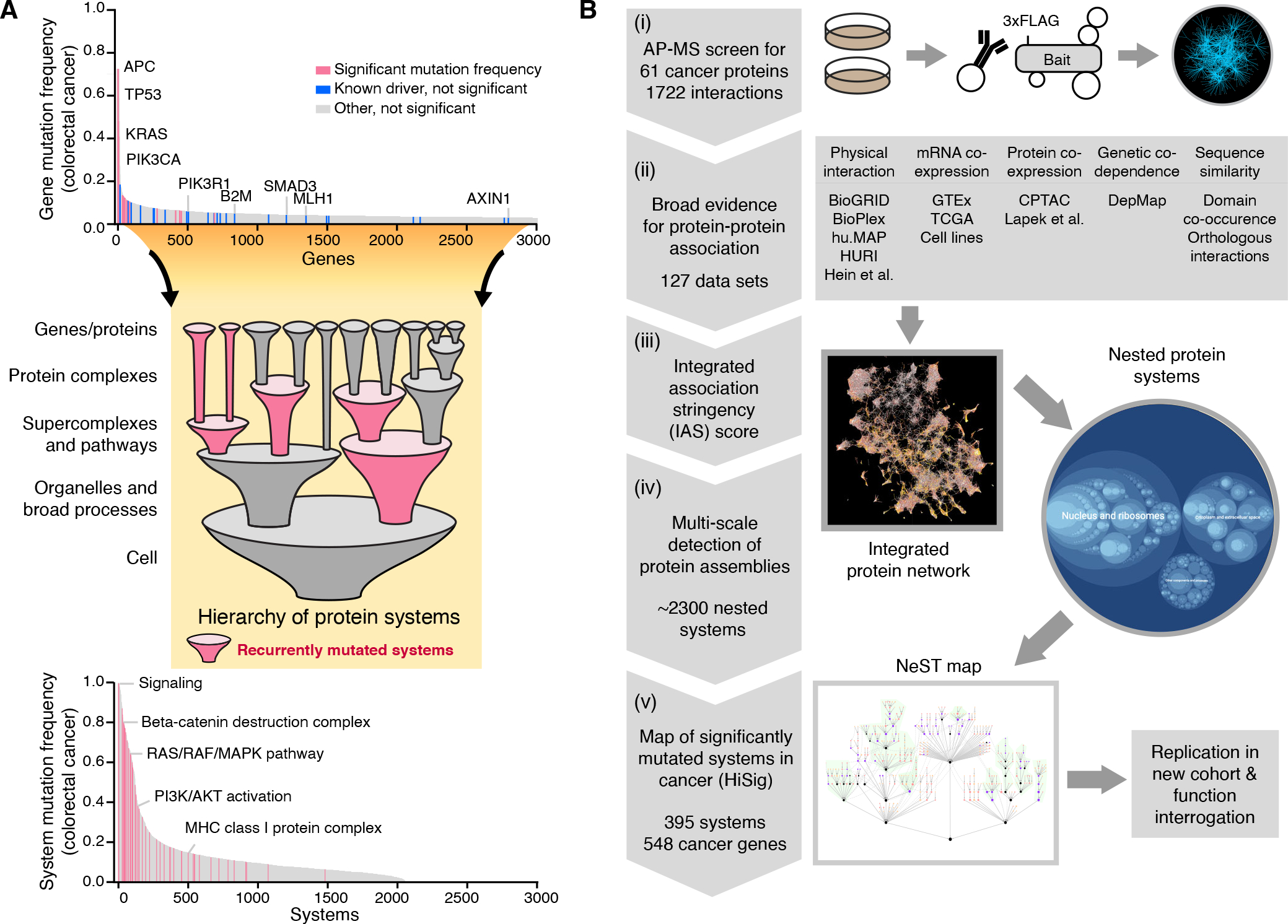
(A) Cancer genes missed by single-gene mutation analysis can be recovered by identification of significantly mutated systems. In the distribution of gene mutation frequencies in colorectal adenocarcinoma (COAD, top), analysis of significantly mutated genes (pink, TCGA pan-cancer atlas) (37) misses a number of known colorectal cancer driver genes (blue, COSMIC Cancer Gene Census) (72). Representative genes from both categories are labeled. When evaluating mutation significance in a hierarchy of protein systems (middle), driver genes missed in the single-gene analysis can be recovered within significantly mutated systems (pink, bottom). (B) Pipeline for assembly of the cancer systems map. (i) Generate cancer protein interactions using affinity purification mass spectrometry (AP-MS). (ii) Collect previous protein association evidence of five major data types. (iii) Integrate all evidence to derive an integrated association stringency (IAS) score network for all pairs of 19,035 human proteins, (iv) Identify a hierarchy of protein systems by multiscale community detection, (v) Identify recurrently mutated systems in the hierarchy by HiSig, defining a cancer systems map, which is validated in independent cohorts and functional studies.
