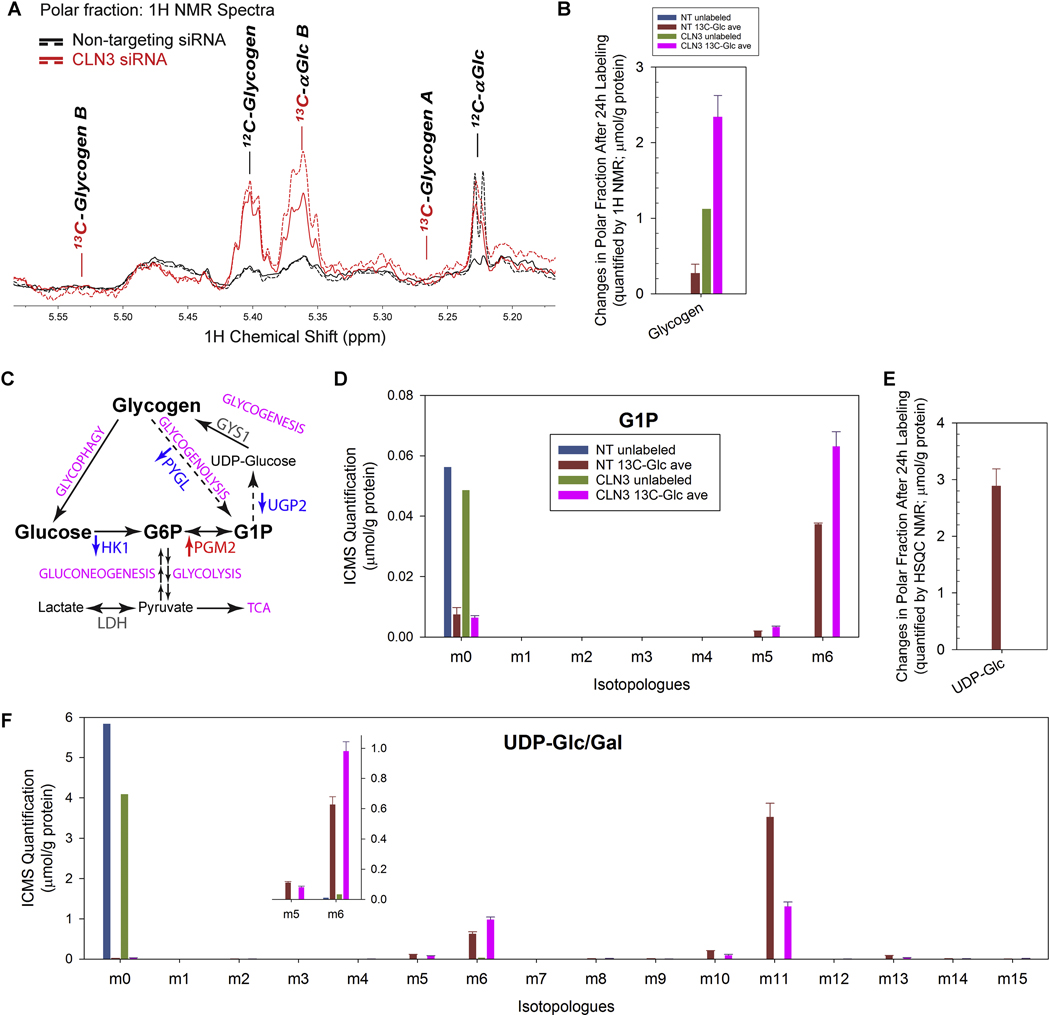
Figure 8. Loss of CLN3 led to glycogen accumulation.
(A) 1H NMR spectra of polar extracts of the cells at 24 h after 13C6-glucose labeling showed that CLN3-deficient RPE-1 cells (in red; solid and dash traces for the duplicated samples) had increased intracellular levels of un-labeled glycogen than non-targeting siRNA-treated control cells (in black; solid and dash traces for the duplicated samples). (B) 1H NMR spectra of polar extracts of the cells at 24 h after 13C6-glucose labeling showed that CLN3-deficient RPE-1 cells had increased intracellular levels of unlabeled glycogen. (C) The glycogen metabolism with enzymes whose transcripts significantly changed (measured by RNA-Seq) upon CLN3 deficiency labeled. Red, up-regulation; blue, down-regulation; grey, unchanged. (D) ICMS analyses of RPE-1 cell polar extracts showed that CLN3 deficiency led to increased levels of intracellular 13C6-glucose-1-phosphate (G1P), the product of glycogen phosphorylase-catalyzed glycogen breakdown, the substrate of UDP-glucose pyro-phosphorylase-catalyzed synthesis of UDP-glucose from G1P and UTP, and a metabolic intermediate that interconverses with G6P. (E) 1H{13C}-HSQC NMR spectra of polar extracts of the cells at 24 h after 13C6-glucose labeling showed that CLN3-deficient RPE-1 cells had depleted intracellular 13C-labeled UDP-glucose (UDP-Glc). (F) ICMS analyses of RPE-1 cell polar extracts showed that CLN3 deficiency led to increased levels of intracellular 13C6-UDP-glucose/galactose (UDP-Glc/Gal; labeling likely at glucose/galactose), but reduced levels of intracellular 13C11-UDP-Glc/Gal (labeling likely at both glucose/galactose and UDP ribose ring) and 13C5-UDP-Glc/Gal (labeling likely at UDP ribose ring). Metabolites amounts were normalized by protein amounts of corresponding samples. Due to limit of resources, the SIRM experiment only included single unlabeled sample and duplicated labeled samples for either non-targeting or CLN3 exon-8 siRNA treatment. For all SIRM quantification results (B, D-F), means and standard errors were plotted for labeled samples.