Figure 2. BET proteins mediate ecDNA hub formation and transcription.
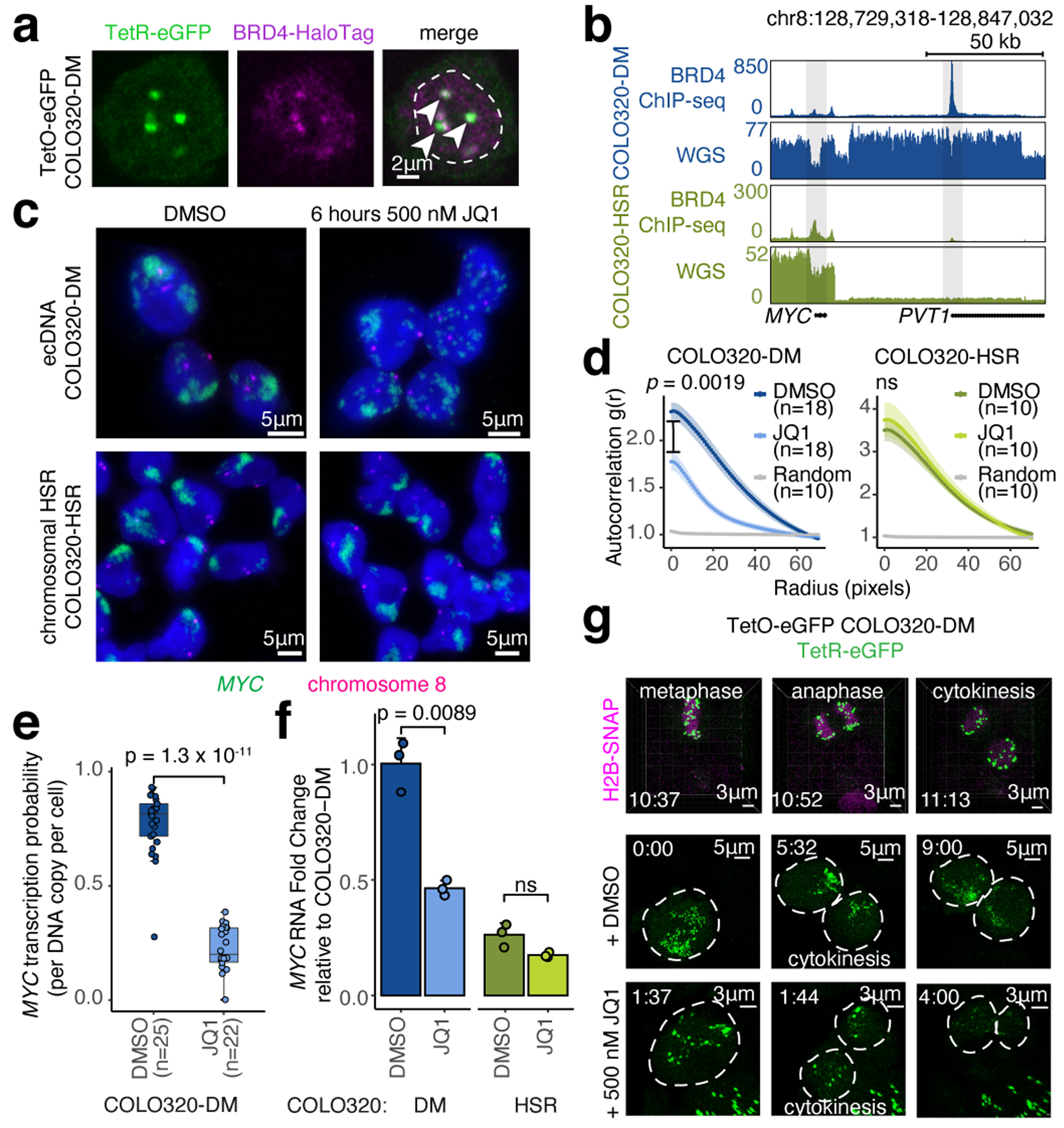
(a) Representative live cell image of ecDNA and BRD4-HaloTag signals in TetO-eGFP COLO320-DM cells (independently repeated twice; dashed line indicates nuclear boundary). (b) BRD4 ChIP-seq and WGS at MYC locus in COLO320-DM and COLO320-HSR cells. (c) Representative DNA FISH images for cells treated with DMSO or 500 nM JQ1 for 6 hours. (d) Clustering measured by autocorrelation g(r) for ecDNAs in COLO320-DM and HSRs in COLO320-HSR treated with DMSO or 500 nM JQ1 for 6 hours. Data are mean ± SEM. P-values determined by two-sided Wilcoxon test at r=0. (e) MYC transcription probability in COLO320-DM treated with DMSO or 500 nM JQ1 for 6 hours (joint DNA/RNA FISH; RNA normalized to ecDNA copy number; box center line, median; box limits, upper and lower quartiles; box whiskers, 1.5x interquartile range). P-values determined by two-sided Wilcoxon test. (f) MYC RNA measured by RT-qPCR for COLO320-DM and COLO320-HSR cells treated either with DMSO or 500 nM JQ1 for 6 hours. Data are mean ± SD between 3 biological replicates. P-values determined by two-sided student’s t-test. (g) Representative live cell images of TetR-eGFP-labeled ecDNAs in TetO-eGFP COLO320-DM cells treated with DMSO or 500 nM JQ1 at indicated timepoints through cell division (independently repeated twice for each condition). H2B-SNAP (top) labels histone H2B in mitotic chromosomes.
