Extended Data Figure 4. Reconstruction of COLO320-DM ecDNA amplicon structure.
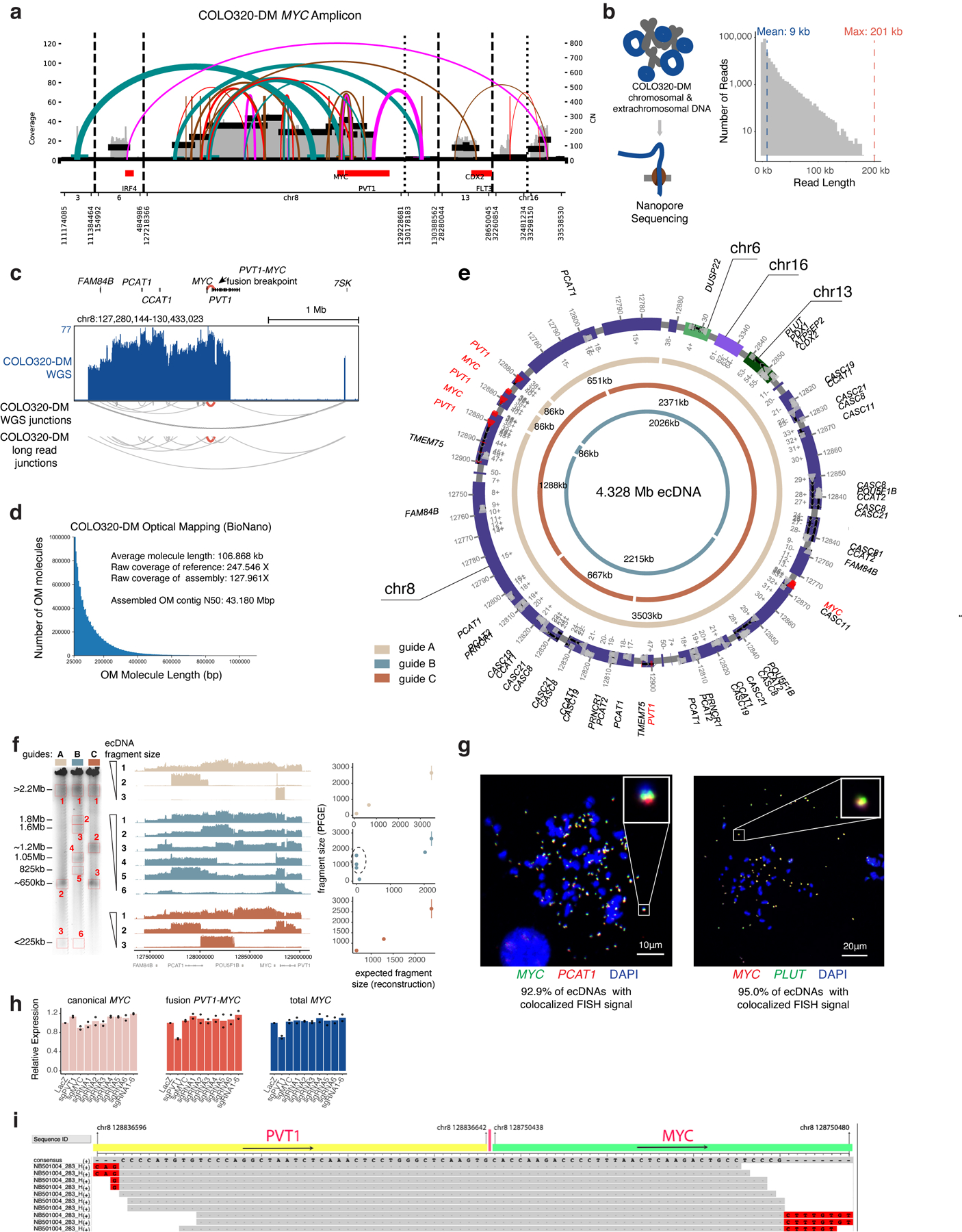
(a) Structural variant (SV) view of AmpliconArchitect (AA) reconstruction of the MYC amplicon in COLO320-DM cells. (b) Nanopore sequencing of COLO320-DM cells (left) and distribution of read lengths. (c) WGS for COLO320-DM with junctions detected by WGS and nanopore sequencing. (d) Molecule lengths used for optical mapping and statistics. (e) Reconstructed COLO320-DM ecDNA after integrating WGS, optical mapping, and in-vitro ecDNA digestion. Chromosomes of origin and corresponding coordinates (hg19) are labeled. Three inner circular tracks (light tan, slate and brown in color; guides A, B and C, respectively) representing expected fragments as a result of Cas9 cleavage using three distinct sgRNAs and their expected sizes. Guide sequences are in Supplementary Table 2 (PFGE_guide_A-C). (f) In-vitro Cas9 digestion of COLO320-DM ecDNA followed by PFGE (left). Fragment sizes were determined based on H. wingei and S. cerevisiae ladders. Uncropped gel image is in Supplementary Figure 1. Middle panel shows short-read sequencing of the MYC ecDNA amplicon for all isolated fragments, ordered by fragment size. Right panel shows concordance of expected fragment sizes by optical mapping reconstruction, and observed fragment sizes by in-vitro Cas9 digestion (discordant fragments circled). Each sgRNA digestion was performed in one independent experiment. (g) Metaphase FISH images showing colocalization of MYC, PCAT1 and PLUT as predicted by optical mapping and in-vitro digestion. N = 20 cells and 1,270 ecDNAs quantified for MYC/PCAT1 DNA FISH and n = 15 cells and 678 ecDNAs for MYC/PLUT DNA FISH from one experiment. (h) RNA expression measured by RT-qPCR for indicated transcripts in COLO320-DM cells stably expressing dCas9-KRAB and indicated sgRNAs (n=2 biological replicates). Canonical MYC was amplified with primers MYC_exon1_fw and MYC_exon2_rv; fusion PVT1-MYC was amplified with PVT1_exon1_fw and MYC_exon2_rv; total MYC was amplified with total_MYC_exon2_fw and total_MYC_exon2_rv. All primer sequences are in Supplementary Table 1 and guide sequences are in Supplementary Table 2. (i) Alignment of junction reads at the PVT1-MYC breakpoint.
