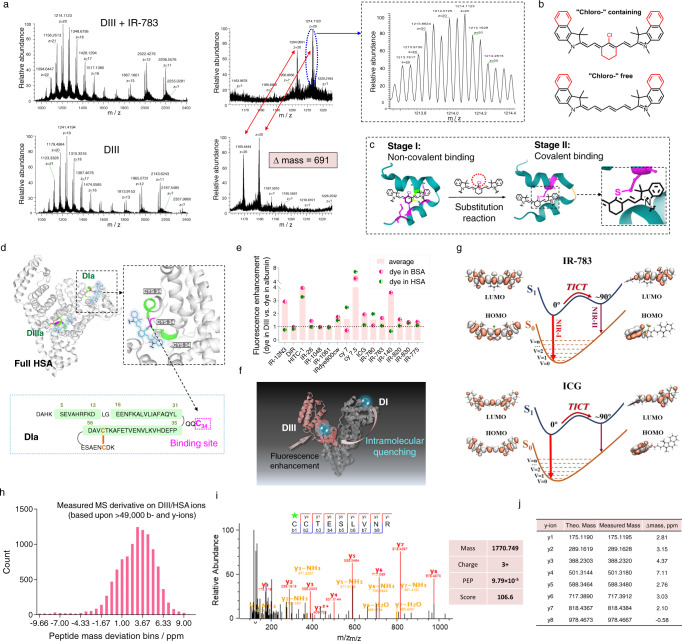
Fig. 3. Mechanism of interaction between cyanine dyes and DIII.
a UHPLC-MS analysis revealed the formation of a covalent bond between DIII and the inserted cyanine dye. The increase in mass was 691 m/z, which was equal to IR-783 (Cl¯ removed). b Core structures of Cl-containing and Cl-free dyes. Some cyanine/polymethine dyes do not contain the red group(s). c Cyanine dyes interact with DIII and its variants in two independent stages. In stage I, the dyes insert into the hydrophobic pocket of DIII and bind to the pocket through non-covalent bonds, resulting in increased brightness. In stage II, covalent bonds stabilize Cl-containing dyes in the pocket. The amino acid residues buried inside the pocket also play essential roles in covalent bond formation. d Full HSA bears two binding sites for cyanine dyes: one site is situated on the surface of DIa (free Cys34 is the only natural cystine containing a free thiol group) and the other is located in the hydrophobic pocket of DIIIa (covalent bonds are formed at Cys475). Lower panel: binding residue in the sequence. e Enhanced fluorescence intensity (1.3–5.9-fold) was recorded for most of the cyanine dyes bound to DIII compared with HSA or BSA. f The fluorescence enhancement is lower for dye@HSA than dye@DIII because the dye bound to DI exhibits intramolecular quenching to the fluorescently enhanced dye bound in the hydrophobic pocket of DIII. g TICT-induced NIR-I and NIR-II states of IR-783 (top) and ICG (bottom) based on DFT and TDDFT calculations. h–j Shotgun proteomics analysis revealed that Cys475 contributes to the formation of the covalent bond. MS/MS spectra (i) and fragment masses (j) of the triply charged tryptic peptide containing the dye-modified cysteine (*indicated) amino acid. The complete y series ions (y1–y8) were acquired using an Orbitrap-based mass analyzer (<0.5 ppm). These ions are labeled on the spectrum and depicted on the sequence (upper panel). The data were analyzed using MaxQuant. Posterior error probability (PEP) and Andromeda scores are presented in the right panel. Source data are provided as a Source Data file.