Fig. 4. Relation of thermal stability and affinity for dsA2/peptide complexes.
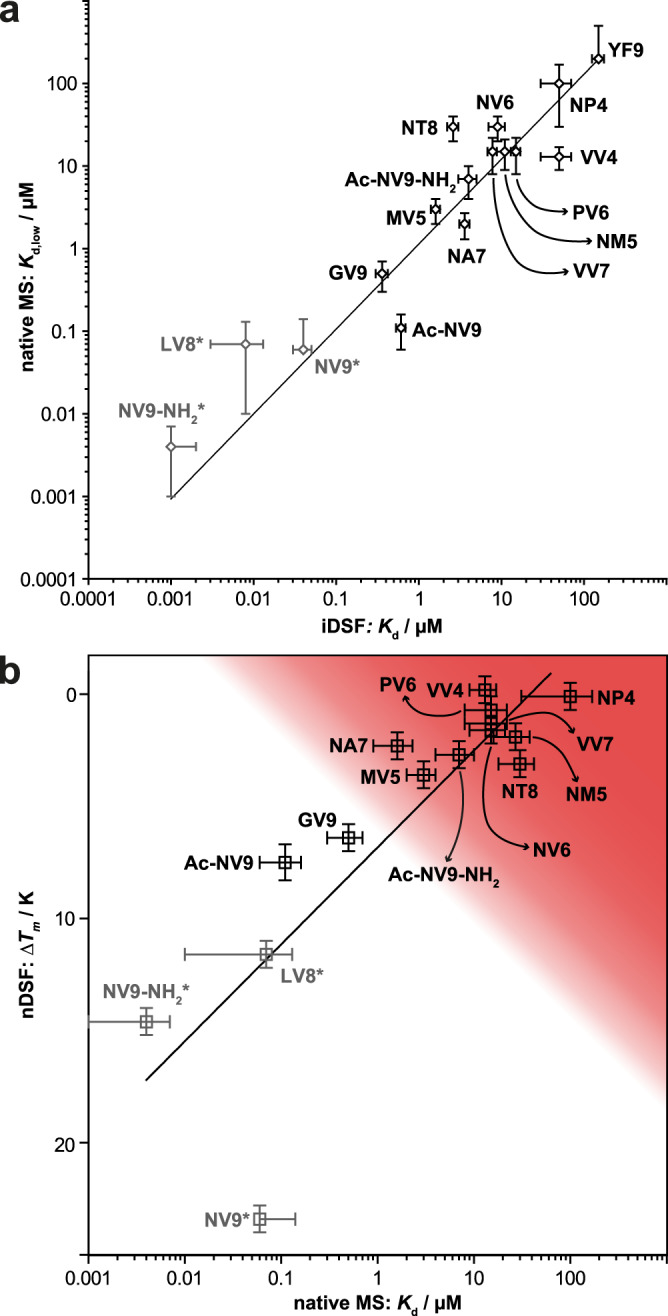
a Affinities determined via iDSF and native MS correlate. Displayed data points represent the relationship of both apparent Kd for all different dsA2/peptide systems analyzed. Both axes are scaled logarithmically. b Thermal denaturation measurements availing intrinsic tryptophans’ change in fluorescence are used to define protein complex stabilization upon peptide binding, whereas an apparent Kd for the various peptides is determined using native MS (10 V acceleration voltage and 150 V cone voltage). The dsA2 complex and peptide are deployed in a ratio of 1:10 (∆Tm) or 1:5 (Kd) depending on the experiment. Peptides showing a small Kd,low for binding dsA2 and being concomitantly able to stabilize the protein complex are defined as strong binders (white area). The remaining peptides (red area) lack crucial features, making them unable to form strong bonds to dsA2 indicated by low binding affinities and melting temperatures. The standard deviation for both methods is displayed by error bars. The x coordinate is displayed logarithmically. *iDSF reaches its limits at affinities below 200 nm, hence the values of grayed-out peptides are not reliable.
